You are browsing environment: FUNGIDB
CAZyme Information: CLCR_08484-t43_1-p1
You are here: Home > Sequence: CLCR_08484-t43_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
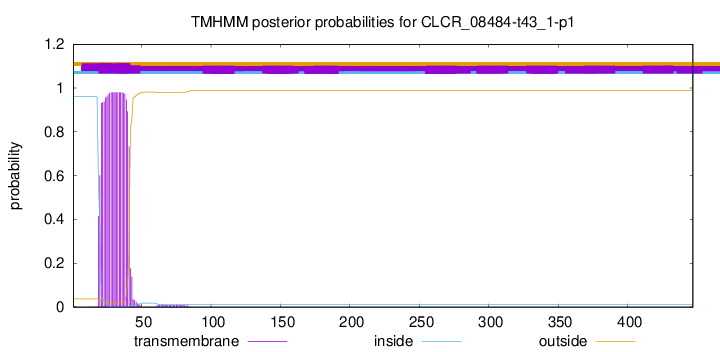
TMHMM annotations
Basic Information help
| Species | Cladophialophora carrionii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Herpotrichiellaceae; Cladophialophora; Cladophialophora carrionii | |||||||||||
| CAZyme ID | CLCR_08484-t43_1-p1 | |||||||||||
| CAZy Family | GT2 | |||||||||||
| CAZyme Description | putative Endo-arabinase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH43 | 66 | 316 | 8.7e-59 | 0.7962264150943397 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 350113 | GH43_ABN-like | 2.58e-69 | 62 | 433 | 3 | 280 | Glycosyl hydrolase family 43 protein such as endo-alpha-L-arabinanase. This glycosyl hydrolase family 43 (GH43) subgroup includes mostly enzymes with alpha-L-arabinofuranosidase (ABF; EC 3.2.1.55) and endo-alpha-L-arabinanase (ABN; EC 3.2.1.99) activities. These are inverting enzymes (i.e. they invert the stereochemistry of the anomeric carbon atom of the substrate) that have an aspartate as the catalytic general base, a glutamate as the catalytic general acid and another aspartate that is responsible for pKa modulation and orienting the catalytic acid. The GH43 ABN enzymes hydrolyze alpha-1,5-L-arabinofuranoside linkages while the ABF enzymes cleave arabinose side chains so that the combined actions of these two enzymes reduce arabinan to L-arabinose and/or arabinooligosaccharides. These arabinan-degrading enzymes are important in the food industry for efficient production of L-arabinose from agricultural waste; L-arabinose is often used as a bioactive sweetener. A common structural feature of GH43 enzymes is a 5-bladed beta-propeller domain that contains the catalytic acid and catalytic base. A long V-shaped groove, partially enclosed at one end, forms a single extended substrate-binding surface across the face of the propeller. |
| 350128 | GH43_ABN-like | 2.26e-18 | 62 | 193 | 3 | 137 | Glycosyl hydrolase family 43 such as arabinan endo-1 5-alpha-L-arabinosidase. This glycosyl hydrolase family 43 (GH43) subgroup includes mostly enzymes with endo-alpha-L-arabinanase (ABN; EC 3.2.1.99) activity. These are inverting enzymes (i.e. they invert the stereochemistry of the anomeric carbon atom of the substrate) that have an aspartate as the catalytic general base, a glutamate as the catalytic general acid and another aspartate that is responsible for pKa modulation and orienting the catalytic acid. The GH43 ABN enzymes hydrolyze alpha-1,5-L-arabinofuranoside linkages. These arabinan-degrading enzymes are important in the food industry for efficient production of L-arabinose from agricultural waste; L-arabinose is often used as a bioactive sweetener. A common structural feature of GH43 enzymes is a 5-bladed beta-propeller domain that contains the catalytic acid and catalytic base. A long V-shaped groove, partially enclosed at one end, forms a single extended substrate-binding surface across the face of the propeller. |
| 350105 | GH43_HoAraf43-like | 6.75e-18 | 68 | 307 | 1 | 203 | Glycosyl hydrolase family 43 protein such as Halothermothrix orenii H 168 alpha-L-arabinofuranosidase (HoAraf43;Hore_20580). This glycosyl hydrolase family 43 (GH43) subgroup includes Halothermothrix orenii H 168 alpha-L-arabinofuranosidase (EC 3.2.1.55) (HoAraf43;Hore_20580). It belongs to the glycosyl hydrolase clan F (according to carbohydrate-active enzymes database (CAZY)) which includes family 43 (GH43) and 62 (GH62) families. This GH43_ HoAraf43-like subgroup includes enzymes that have been annotated as having xylan-digesting beta-xylosidase (EC 3.2.1.37) and xylanase (endo-alpha-L-arabinanase, EC 3.2.1.8) activities. GH43 are inverting enzymes (i.e. they invert the stereochemistry of the anomeric carbon atom of the substrate) that have an aspartate as the catalytic general base, a glutamate as the catalytic general acid and another aspartate that is responsible for pKa modulation and orienting the catalytic acid. Many GH43 enzymes display both alpha-L-arabinofuranosidase and beta-D-xylosidase activity using aryl-glycosides as substrates. A common structural feature of GH43 enzymes is a 5-bladed beta-propeller domain that contains the catalytic acid and catalytic base. A long V-shaped groove, partially enclosed at one end, forms a single extended substrate-binding surface across the face of the propeller. |
| 350102 | GH43_ABN | 4.84e-13 | 68 | 295 | 1 | 204 | Glycosyl hydrolase family 43. This glycosyl hydrolase family 43 (GH43) subgroup includes mostly enzymes with alpha-L-arabinofuranosidase (ABF; EC 3.2.1.55) and endo-alpha-L-arabinanase (ABN; EC 3.2.1.99) activities. These are inverting enzymes (i.e. they invert the stereochemistry of the anomeric carbon atom of the substrate) that have an aspartate as the catalytic general base, a glutamate as the catalytic general acid and another aspartate that is responsible for pKa modulation and orienting the catalytic acid. The GH43 ABN enzymes hydrolyze alpha-1,5-L-arabinofuranoside linkages while the ABF enzymes cleave arabinose side chains so that the combined actions of these two enzymes reduce arabinan to L-arabinose and/or arabinooligosaccharides. These arabinan-degrading enzymes are important in the food industry for efficient production of L-arabinose from agricultural waste; L-arabinose is often used as a bioactive sweetener. A common structural feature of GH43 enzymes is a 5-bladed beta-propeller domain that contains the catalytic acid and catalytic base. A long V-shaped groove, partially enclosed at one end, forms a single extended substrate-binding surface across the face of the propeller. |
| 398349 | Glyco_hydro_43 | 5.09e-12 | 62 | 296 | 5 | 205 | Glycosyl hydrolases family 43. The glycosyl hydrolase family 43 contains members that are arabinanases. Arabinanases hydrolyze the alpha-1,5-linked L-arabinofuranoside backbone of plant cell wall arabinans. The structure of arabinanase Arb43A from Cellvibrio japonicus reveals a five-bladed beta-propeller fold. A long V-shaped groove, partially enclosed at one end, forms a single extended substrate-binding surface across the face of the propeller. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.35e-45 | 63 | 345 | 17 | 267 | |
| 1.53e-45 | 65 | 309 | 46 | 273 | |
| 2.19e-44 | 63 | 314 | 35 | 262 | |
| 2.19e-44 | 63 | 314 | 35 | 262 | |
| 2.19e-44 | 63 | 314 | 35 | 262 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.95e-12 | 56 | 169 | 26 | 138 | Crystal Structure of Endo-1,5-alpha-L-arabinanase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],4KC7_B Crystal Structure of Endo-1,5-alpha-L-arabinanase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],4KC7_C Crystal Structure of Endo-1,5-alpha-L-arabinanase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],4KC8_A Crystal Structure of Endo-1,5-alpha-L-arabinanase from Thermotoga petrophila RKU-1 in complex with TRIS [Thermotoga petrophila RKU-1],4KC8_B Crystal Structure of Endo-1,5-alpha-L-arabinanase from Thermotoga petrophila RKU-1 in complex with TRIS [Thermotoga petrophila RKU-1],4KC8_C Crystal Structure of Endo-1,5-alpha-L-arabinanase from Thermotoga petrophila RKU-1 in complex with TRIS [Thermotoga petrophila RKU-1] |
|
| 2.33e-07 | 69 | 169 | 15 | 117 | Crystal structure of the glycoside hydrolase, family 43 YxiA protein from Bacillus licheniformis. Northeast Structural Genomics Consortium Target BiR14. [Bacillus licheniformis DSM 13 = ATCC 14580],3LV4_B Crystal structure of the glycoside hydrolase, family 43 YxiA protein from Bacillus licheniformis. Northeast Structural Genomics Consortium Target BiR14. [Bacillus licheniformis DSM 13 = ATCC 14580] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.51e-11 | 56 | 169 | 23 | 135 | Extracellular endo-alpha-(1->5)-L-arabinanase OS=Thermotoga petrophila (strain ATCC BAA-488 / DSM 13995 / JCM 10881 / RKU-1) OX=390874 GN=Tpet_0637 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
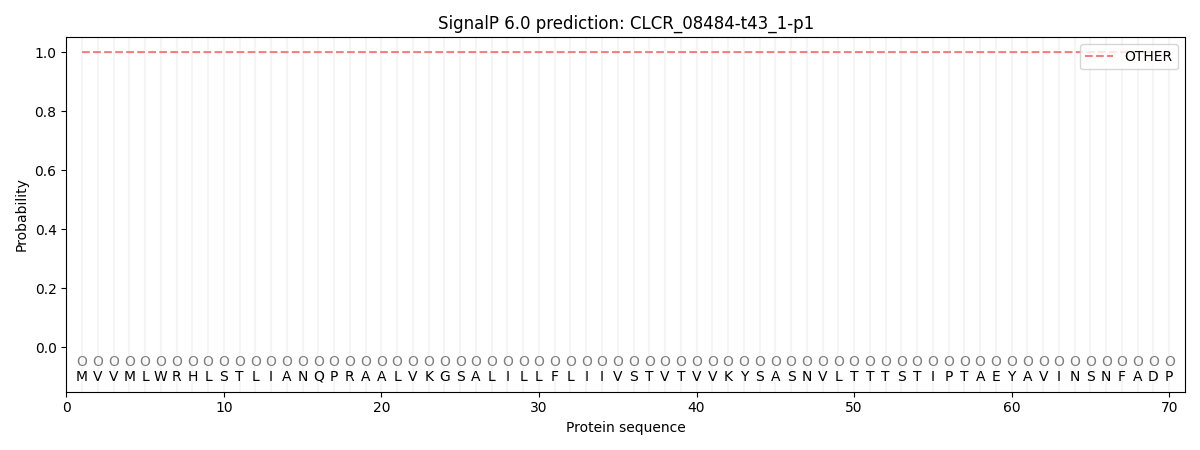
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000021 | 0.000009 |