You are browsing environment: FUNGIDB
CAZyme Information: CLCR_06851-t43_1-p1
You are here: Home > Sequence: CLCR_06851-t43_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Cladophialophora carrionii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Herpotrichiellaceae; Cladophialophora; Cladophialophora carrionii | |||||||||||
| CAZyme ID | CLCR_06851-t43_1-p1 | |||||||||||
| CAZy Family | GH76 | |||||||||||
| CAZyme Description | Endo-1,6-beta-D-glucanase BGN16.3 | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.75:6 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH30 | 97 | 534 | 1e-94 | 0.9784172661870504 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 227807 | XynC | 1.92e-27 | 58 | 543 | 16 | 425 | O-Glycosyl hydrolase [Cell wall/membrane/envelope biogenesis]. |
| 307945 | Glyco_hydro_30 | 1.38e-21 | 111 | 394 | 3 | 273 | Glycosyl hydrolase family 30 TIM-barrel domain. |
| 407314 | Glyco_hydro_30C | 0.010 | 480 | 534 | 11 | 63 | Glycosyl hydrolase family 30 beta sandwich domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.15e-220 | 16 | 541 | 13 | 513 | |
| 7.96e-97 | 82 | 538 | 51 | 497 | |
| 7.96e-97 | 82 | 538 | 51 | 497 | |
| 7.50e-94 | 82 | 538 | 42 | 487 | |
| 7.50e-94 | 82 | 538 | 42 | 487 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.17e-27 | 101 | 513 | 69 | 470 | The endo-beta1,6-glucanase BT3312 [Bacteroides thetaiotaomicron],5NGK_B The endo-beta1,6-glucanase BT3312 [Bacteroides thetaiotaomicron],5NGK_C The endo-beta1,6-glucanase BT3312 [Bacteroides thetaiotaomicron],5NGL_A The endo-beta1,6-glucanase BT3312 [Bacteroides thetaiotaomicron],5NGL_B The endo-beta1,6-glucanase BT3312 [Bacteroides thetaiotaomicron],5NGL_C The endo-beta1,6-glucanase BT3312 [Bacteroides thetaiotaomicron] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.33e-94 | 82 | 538 | 42 | 487 | Endo-1,6-beta-D-glucanase neg1 OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=neg1 PE=1 SV=1 |
|
| 1.99e-94 | 100 | 538 | 69 | 489 | Endo-1,6-beta-D-glucanase BGN16.3 OS=Trichoderma harzianum OX=5544 PE=1 SV=1 |
|
| 3.40e-88 | 47 | 540 | 5 | 479 | Endo-1,6-beta-D-glucanase OS=Neurospora crassa (strain ATCC 24698 / 74-OR23-1A / CBS 708.71 / DSM 1257 / FGSC 987) OX=367110 GN=neg-1 PE=1 SV=2 |
|
| 2.03e-21 | 88 | 533 | 79 | 515 | Putative glucosylceramidase 4 OS=Caenorhabditis elegans OX=6239 GN=gba-4 PE=3 SV=2 |
|
| 3.55e-19 | 107 | 465 | 102 | 455 | Putative glucosylceramidase 3 OS=Caenorhabditis elegans OX=6239 GN=gba-3 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
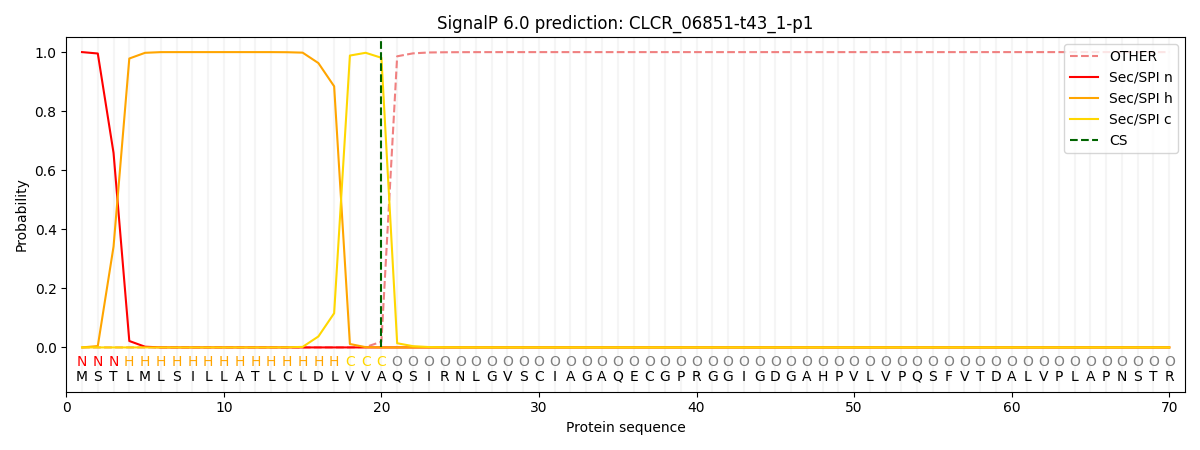
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000224 | 0.999750 | CS pos: 20-21. Pr: 0.9807 |
