You are browsing environment: FUNGIDB
CAZyme Information: CKF44_01214-t42_1-p1
You are here: Home > Sequence: CKF44_01214-t42_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
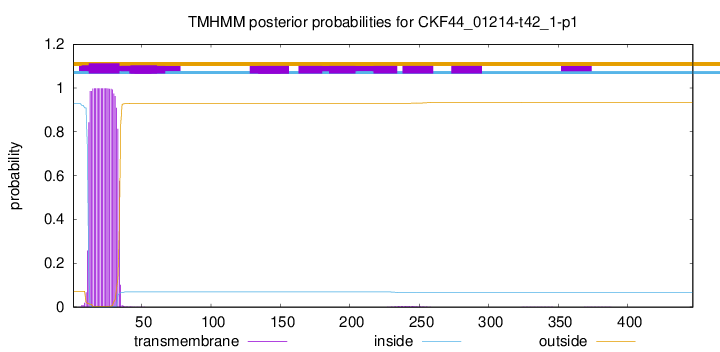
TMHMM annotations
Basic Information help
| Species | Cryptococcus neoformans | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Arthropoda; Insecta; ; Eriococcidae; Cryptococcus; Cryptococcus neoformans | |||||||||||
| CAZyme ID | CKF44_01214-t42_1-p1 | |||||||||||
| CAZy Family | GT90 | |||||||||||
| CAZyme Description | alpha-1,6-mannosyltransferase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 226297 | OCH1 | 3.91e-14 | 120 | 338 | 47 | 236 | Mannosyltransferase OCH1 or related enzyme [Cell wall/membrane/envelope biogenesis]. |
| 398274 | Gly_transf_sug | 6.30e-10 | 173 | 257 | 7 | 86 | Glycosyltransferase sugar-binding region containing DXD motif. The DXD motif is a short conserved motif found in many families of glycosyltransferases, which add a range of different sugars to other sugars, phosphates and proteins. DXD-containing glycosyltransferases all use nucleoside diphosphate sugars as donors and require divalent cations, usually manganese. The DXD motif is expected to play a carbohydrate binding role in sugar-nucleoside diphosphate and manganese dependent glycosyltransferases. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 447 | 1 | 447 | |
| 0.0 | 1 | 447 | 1 | 447 | |
| 1.15e-258 | 1 | 445 | 1 | 438 | |
| 3.40e-258 | 1 | 445 | 1 | 439 | |
| 9.47e-252 | 1 | 377 | 1 | 395 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 9.26e-28 | 114 | 441 | 58 | 376 | Initiation-specific alpha-1,6-mannosyltransferase OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=OCH1 PE=3 SV=1 |
|
| 1.82e-26 | 155 | 441 | 142 | 394 | Initiation-specific alpha-1,6-mannosyltransferase OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=och1 PE=1 SV=2 |
|
| 4.93e-19 | 225 | 418 | 196 | 362 | Putative glycosyltransferase HOC1 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=HOC1 PE=1 SV=3 |
|
| 6.73e-17 | 150 | 445 | 91 | 474 | Initiation-specific alpha-1,6-mannosyltransferase OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=OCH1 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.999933 | 0.000113 |