You are browsing environment: FUNGIDB
CAZyme Information: CIMG_02738-t26_1-p1
You are here: Home > Sequence: CIMG_02738-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Coccidioides immitis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Onygenaceae; Coccidioides; Coccidioides immitis | |||||||||||
| CAZyme ID | CIMG_02738-t26_1-p1 | |||||||||||
| CAZy Family | GH13 | |||||||||||
| CAZyme Description | polysaccharide synthase Cps1 | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT2 | 45 | 284 | 1.9e-36 | 0.9826086956521739 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 224055 | MdlB | 5.20e-93 | 473 | 1009 | 21 | 566 | ABC-type multidrug transport system, ATPase and permease component [Defense mechanisms]. |
| 133056 | GT2_HAS | 4.20e-92 | 45 | 289 | 1 | 235 | Hyaluronan synthases catalyze polymerization of hyaluronan. Hyaluronan synthases (HASs) are bi-functional glycosyltransferases that catalyze polymerization of hyaluronan. HASs transfer both GlcUA and GlcNAc in beta-(1,3) and beta-(1,4) linkages, respectively to the hyaluronan chain using UDP-GlcNAc and UDP-GlcUA as substrates. HA is made as a free glycan, not attached to a protein or lipid. HASs do not need a primer for HA synthesis; they initiate HA biosynthesis de novo with only UDP-GlcNAc, UDP-GlcUA, and Mg2+. Hyaluronan (HA) is a linear heteropolysaccharide composed of (1-3)-linked beta-D-GlcUA-beta-D-GlcNAc disaccharide repeats. It can be found in vertebrates and a few microbes and is typically on the cell surface or in the extracellular space, but is also found inside mammalian cells. Hyaluronan has several physiochemical and biological functions such as space filling, lubrication, and providing a hydrated matrix through which cells can migrate. |
| 213220 | ABCC_ATM1_transporter | 6.61e-85 | 777 | 1006 | 1 | 235 | ATP-binding cassette domain of iron-sulfur clusters transporter, subfamily C. ATM1 is an ABC transporter that is expressed in the mitochondria. Although the specific function of ATM1 is unknown, its disruption results in the accumulation of excess mitochondrial iron, loss of mitochondrial cytochromes, oxidative damage to mitochondrial DNA, and decreased levels of cytosolic heme proteins. ABC transporters are a large family of proteins involved in the transport of a wide variety of different compounds, like sugars, ions, peptides, and more complex organic molecules. The nucleotide binding domain shows the highest similarity between all members of the family. ABC transporters are a subset of nucleotide hydrolases that contain a signature motif, Q-loop, and H-loop/switch region, in addition to, the Walker A motif/P-loop and Walker B motif commonly found in a number of ATP- and GTP-binding and hydrolyzing proteins. |
| 213216 | ABC_MTABC3_MDL1_MDL2 | 2.65e-80 | 777 | 1005 | 1 | 236 | ATP-binding cassette domain of a mitochondrial protein MTABC3 and related proteins. MTABC3 (also known as ABCB6) is a mitochondrial ATP-binding cassette protein involved in iron homeostasis and one of four ABC transporters expressed in the mitochondrial inner membrane, the other three being MDL1(ABC7), MDL2, and ATM1. In fact, the yeast MDL1 (multidrug resistance-like protein 1) and MDL2 (multidrug resistance-like protein 2) transporters are also included in this CD. MDL1 is an ATP-dependent permease that acts as a high-copy suppressor of ATM1 and is thought to have a role in resistance to oxidative stress. Interestingly, subfamily B is more closely related to the carboxyl-terminal component of subfamily C than the two halves of ABCC molecules are with one another. |
| 213218 | ABCC_MsbA | 1.08e-78 | 777 | 1004 | 1 | 234 | ATP-binding cassette domain of the bacterial lipid flippase and related proteins, subfamily C. MsbA is an essential ABC transporter, closely related to eukaryotic MDR proteins. ABC transporters are a large family of proteins involved in the transport of a wide variety of different compounds, like sugars, ions, peptides, and more complex organic molecules. The nucleotide binding domain shows the highest similarity between all members of the family. ABC transporters are a subset of nucleotide hydrolases that contain a signature motif, Q-loop, and H-loop/switch region, in addition to, the Walker A motif/P-loop and Walker B motif commonly found in a number of ATP- and GTP-binding and hydrolyzing proteins. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 6.50e-229 | 9 | 1004 | 9 | 997 | |
| 9.67e-140 | 1 | 414 | 1 | 418 | |
| 3.78e-139 | 1 | 414 | 1 | 418 | |
| 6.92e-139 | 1 | 422 | 1 | 426 | |
| 5.79e-138 | 1 | 414 | 1 | 418 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.30e-48 | 449 | 1004 | 19 | 593 | Structure of a bacterial Atm1-family ABC exporter with MgAMPPNP bound [Novosphingobium aromaticivorans DSM 12444],6PAR_B Structure of a bacterial Atm1-family ABC exporter with MgAMPPNP bound [Novosphingobium aromaticivorans DSM 12444],6PAR_C Structure of a bacterial Atm1-family ABC exporter with MgAMPPNP bound [Novosphingobium aromaticivorans DSM 12444],6PAR_D Structure of a bacterial Atm1-family ABC exporter with MgAMPPNP bound [Novosphingobium aromaticivorans DSM 12444],6PAR_E Structure of a bacterial Atm1-family ABC exporter with MgAMPPNP bound [Novosphingobium aromaticivorans DSM 12444],6PAR_F Structure of a bacterial Atm1-family ABC exporter with MgAMPPNP bound [Novosphingobium aromaticivorans DSM 12444],6VQT_A Structure of a bacterial Atm1-family ABC exporter with MgADPVO4 bound [Novosphingobium aromaticivorans DSM 12444],6VQT_B Structure of a bacterial Atm1-family ABC exporter with MgADPVO4 bound [Novosphingobium aromaticivorans DSM 12444],6VQU_A Structure of a bacterial Atm1-family ABC exporter [Novosphingobium aromaticivorans DSM 12444],6VQU_B Structure of a bacterial Atm1-family ABC exporter [Novosphingobium aromaticivorans DSM 12444] |
|
| 1.75e-48 | 449 | 1004 | 19 | 593 | Structure of a bacterial Atm1-family ABC transporter with MgADP bound [Novosphingobium aromaticivorans DSM 12444],6PAM_B Structure of a bacterial Atm1-family ABC transporter with MgADP bound [Novosphingobium aromaticivorans DSM 12444],6PAM_C Structure of a bacterial Atm1-family ABC transporter with MgADP bound [Novosphingobium aromaticivorans DSM 12444],6PAM_D Structure of a bacterial Atm1-family ABC transporter with MgADP bound [Novosphingobium aromaticivorans DSM 12444],6PAM_E Structure of a bacterial Atm1-family ABC transporter with MgADP bound [Novosphingobium aromaticivorans DSM 12444],6PAM_F Structure of a bacterial Atm1-family ABC transporter with MgADP bound [Novosphingobium aromaticivorans DSM 12444],6PAM_G Structure of a bacterial Atm1-family ABC transporter with MgADP bound [Novosphingobium aromaticivorans DSM 12444],6PAM_H Structure of a bacterial Atm1-family ABC transporter with MgADP bound [Novosphingobium aromaticivorans DSM 12444] |
|
| 4.54e-48 | 726 | 1003 | 327 | 608 | Chain U, Bile salt export pump [Homo sapiens] |
|
| 4.80e-48 | 726 | 1003 | 372 | 653 | Chain U, Bile salt export pump [Homo sapiens],7E1A_U Chain U, Bile salt export pump [Homo sapiens] |
|
| 7.78e-48 | 449 | 1004 | 19 | 593 | Structure of a bacterial Atm1-family ABC exporter with ATP bound [Novosphingobium aromaticivorans DSM 12444],6PAO_B Structure of a bacterial Atm1-family ABC exporter with ATP bound [Novosphingobium aromaticivorans DSM 12444] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.13e-98 | 3 | 413 | 30 | 438 | Type 2 glycosyltransferase OS=Magnaporthe oryzae (strain 70-15 / ATCC MYA-4617 / FGSC 8958) OX=242507 GN=GT2 PE=1 SV=1 |
|
| 1.19e-91 | 1 | 405 | 1 | 401 | Type 2 glycosyltransferase OS=Zymoseptoria tritici (strain CBS 115943 / IPO323) OX=336722 GN=GT2 PE=3 SV=1 |
|
| 2.89e-86 | 2 | 440 | 33 | 473 | Type 2 glycosyltransferase OS=Gibberella zeae (strain ATCC MYA-4620 / CBS 123657 / FGSC 9075 / NRRL 31084 / PH-1) OX=229533 GN=GT2 PE=2 SV=1 |
|
| 2.24e-64 | 405 | 1004 | 198 | 817 | Heavy metal tolerance protein OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=hmt1 PE=2 SV=3 |
|
| 5.69e-55 | 443 | 1008 | 209 | 789 | ABC transporter aclQ OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=aclQ PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.995488 | 0.004522 |
TMHMM Annotations download full data without filtering help
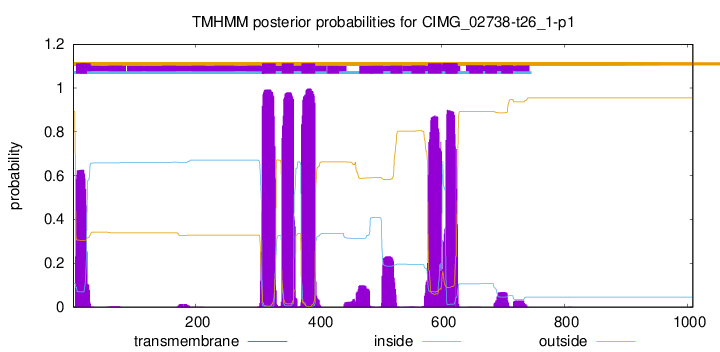
| Start | End |
|---|---|
| 5 | 22 |
| 308 | 330 |
| 340 | 359 |
| 372 | 394 |
| 578 | 600 |
| 607 | 626 |
