You are browsing environment: FUNGIDB
CAZyme Information: CIHG_01964-t26_1-p1
You are here: Home > Sequence: CIHG_01964-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Coccidioides immitis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Onygenaceae; Coccidioides; Coccidioides immitis | |||||||||||
| CAZyme ID | CIHG_01964-t26_1-p1 | |||||||||||
| CAZy Family | CBM21 | |||||||||||
| CAZyme Description | cell wall alpha-1,3-glucan synthase ags1 | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 2.4.1.183:36 | 2.4.1.-:11 |
|---|
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 235490 | PRK05478 | 0.0 | 8 | 480 | 2 | 465 | 3-isopropylmalate dehydratase large subunit. |
| 153133 | IPMI | 0.0 | 35 | 474 | 1 | 382 | 3-isopropylmalate dehydratase catalyzes the isomerization between 2-isopropylmalate and 3-isopropylmalate. Aconatase-like catalytic domain of 3-isopropylmalate dehydratase and related uncharacterized proteins. 3-isopropylmalate dehydratase catalyzes the isomerization between 2-isopropylmalate and 3-isopropylmalate, via the formation of 2-isopropylmaleate 3-isopropylmalate. IPMI is involved in fungal and bacterial leucine biosynthesis and is also found in eukaryotes. |
| 183543 | PRK12466 | 0.0 | 7 | 481 | 2 | 468 | 3-isopropylmalate dehydratase large subunit. |
| 272940 | leuC | 0.0 | 8 | 480 | 2 | 465 | 3-isopropylmalate dehydratase, large subunit. Members of this family are 3-isopropylmalate dehydratase, large subunit, or the large subunit domain of single-chain forms. Homoaconitase, aconitase, and 3-isopropylmalate dehydratase have similar overall structures. All are dehydratases (EC 4.2.1.-) and bind a Fe-4S iron-sulfur cluster. 3-isopropylmalate dehydratase is split into large (leuC) and small (leuD) chains in eubacteria. Several pairs of archaeal proteins resemble the leuC and leuD pair in length and sequence but even more closely resemble the respective domains of homoaconitase, and their identity is uncertain. These homologs are now described by a separate model of subfamily (rather than equivalog) homology type, and the priors and cutoffs for this model have been changed to focus this equivalog family more narrowly. [Amino acid biosynthesis, Pyruvate family] |
| 223143 | LeuC | 0.0 | 7 | 480 | 1 | 422 | Homoaconitase/3-isopropylmalate dehydratase large subunit [Amino acid transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 901 | 2644 | 21 | 2245 | |
| 0.0 | 901 | 2644 | 21 | 2245 | |
| 0.0 | 901 | 2644 | 21 | 2249 | |
| 0.0 | 894 | 2646 | 14 | 2258 | |
| 0.0 | 901 | 2643 | 20 | 2259 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.23e-69 | 29 | 480 | 44 | 443 | The reduced form of MJ0499 [Methanocaldococcus jannaschii DSM 2661],4NQY_B The reduced form of MJ0499 [Methanocaldococcus jannaschii DSM 2661] |
|
| 1.66e-65 | 33 | 480 | 50 | 445 | Crystal structure of IPM isomerase large subunit from methanococcus jannaschii (MJ0499) [Methanocaldococcus jannaschii DSM 2661] |
|
| 1.45e-63 | 10 | 479 | 23 | 441 | Crystal structure of homoaconitase large subunit from methanococcus jannaschii (MJ1003) [Methanocaldococcus jannaschii DSM 2661],4KP2_B Crystal structure of homoaconitase large subunit from methanococcus jannaschii (MJ1003) [Methanocaldococcus jannaschii DSM 2661] |
|
| 2.34e-27 | 7 | 619 | 34 | 647 | STRUCTURE OF ACTIVATED ACONITASE. FORMATION OF THE (4FE-4S) CLUSTER IN THE CRYSTAL [Sus scrofa],6ACN_A STRUCTURE OF ACTIVATED ACONITASE. FORMATION OF THE (4FE-4S) CLUSTER IN THE CRYSTAL [Sus scrofa],7ACN_A CRYSTAL STRUCTURES OF ACONITASE WITH ISOCITRATE AND NITROISOCITRATE BOUND [Sus scrofa] |
|
| 5.35e-27 | 7 | 619 | 33 | 646 | Chain A, PROTEIN (ACONITASE) [Sus scrofa],1C97_A S642A:ISOCITRATE COMPLEX OF ACONITASE [Bos taurus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 0.0 | 893 | 2628 | 14 | 2168 | Cell wall alpha-1,3-glucan synthase mok13 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=mok13 PE=3 SV=2 |
|
| 0.0 | 900 | 2645 | 22 | 2211 | Cell wall alpha-1,3-glucan synthase mok11 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=mok11 PE=3 SV=2 |
|
| 0.0 | 902 | 2645 | 28 | 2224 | Cell wall alpha-1,3-glucan synthase ags1 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=ags1 PE=1 SV=3 |
|
| 0.0 | 991 | 2644 | 339 | 2181 | Cell wall alpha-1,3-glucan synthase mok12 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=mok12 PE=3 SV=1 |
|
| 3.10e-303 | 9 | 735 | 3 | 733 | 3-isopropylmalate dehydratase OS=Rhizomucor pusillus OX=4840 GN=LEUA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000055 | 0.000000 |
TMHMM Annotations download full data without filtering help
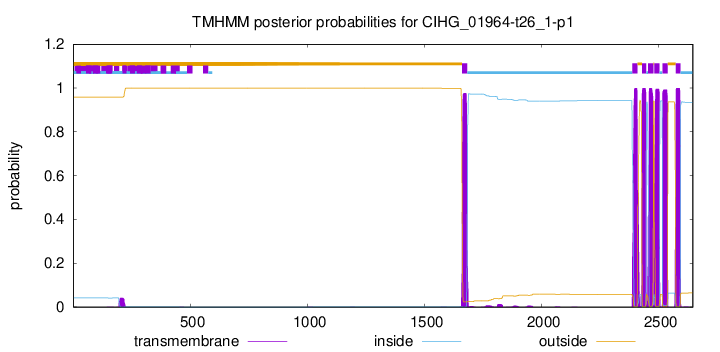
| Start | End |
|---|---|
| 1660 | 1682 |
| 2387 | 2409 |
| 2429 | 2446 |
| 2455 | 2477 |
| 2481 | 2503 |
| 2516 | 2538 |
| 2573 | 2592 |
