You are browsing environment: FUNGIDB
CAZyme Information: CGB_B1360W-t26_1-p1
You are here: Home > Sequence: CGB_B1360W-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
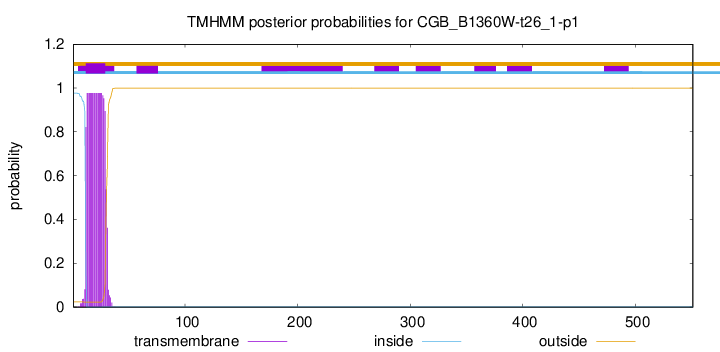
TMHMM annotations
Basic Information help
| Species | Cryptococcus gattii VGI | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Arthropoda; Insecta; ; Eriococcidae; Cryptococcus; Cryptococcus gattii VGI | |||||||||||
| CAZyme ID | CGB_B1360W-t26_1-p1 | |||||||||||
| CAZy Family | CBM67|GH78 | |||||||||||
| CAZyme Description | capsule structure designer protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE17 | 121 | 291 | 8.6e-20 | 0.9818181818181818 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 238141 | SGNH_hydrolase | 2.39e-15 | 118 | 301 | 1 | 187 | SGNH_hydrolase, or GDSL_hydrolase, is a diverse family of lipases and esterases. The tertiary fold of the enzyme is substantially different from that of the alpha/beta hydrolase family and unique among all known hydrolases; its active site closely resembles the typical Ser-His-Asp(Glu) triad from other serine hydrolases, but may lack the carboxlic acid. |
| 404371 | Lipase_GDSL_2 | 0.001 | 120 | 292 | 1 | 175 | GDSL-like Lipase/Acylhydrolase family. This family of presumed lipases and related enzymes are similar to pfam00657. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 9.81e-25 | 79 | 530 | 712 | 1125 | |
| 9.81e-25 | 85 | 530 | 718 | 1125 | |
| 1.29e-16 | 93 | 316 | 351 | 573 | |
| 3.53e-14 | 98 | 315 | 15 | 226 | |
| 3.73e-14 | 98 | 305 | 15 | 213 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.16e-12 | 102 | 312 | 19 | 220 | Crystal structure of a two-domain esterase (CEX) active on acetylated mannans co-crystallized with mannopentaose [Roseburia intestinalis L1-82],6HH9_B Crystal structure of a two-domain esterase (CEX) active on acetylated mannans co-crystallized with mannopentaose [Roseburia intestinalis L1-82],6HH9_C Crystal structure of a two-domain esterase (CEX) active on acetylated mannans co-crystallized with mannopentaose [Roseburia intestinalis L1-82],6HH9_D Crystal structure of a two-domain esterase (CEX) active on acetylated mannans co-crystallized with mannopentaose [Roseburia intestinalis L1-82] |
|
| 1.29e-10 | 102 | 312 | 19 | 220 | Crystal structure of a two-domain esterase (CEX) active on acetylated mannans [Roseburia intestinalis L1-82],6HFZ_B Crystal structure of a two-domain esterase (CEX) active on acetylated mannans [Roseburia intestinalis L1-82],6HFZ_C Crystal structure of a two-domain esterase (CEX) active on acetylated mannans [Roseburia intestinalis L1-82],6HFZ_D Crystal structure of a two-domain esterase (CEX) active on acetylated mannans [Roseburia intestinalis L1-82] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as OTHER
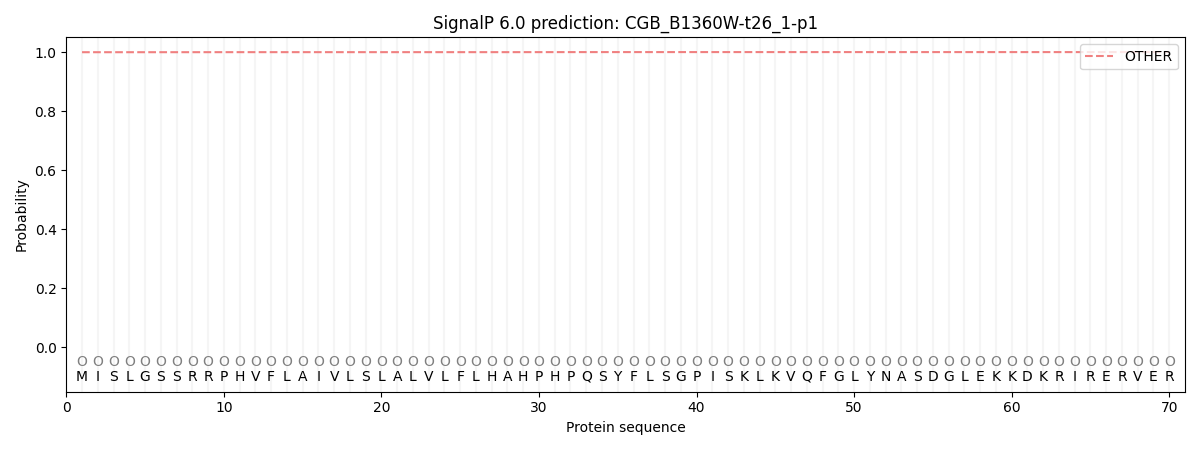
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.999677 | 0.000371 |