You are browsing environment: FUNGIDB
CAZyme Information: CDH60700.1
You are here: Home > Sequence: CDH60700.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
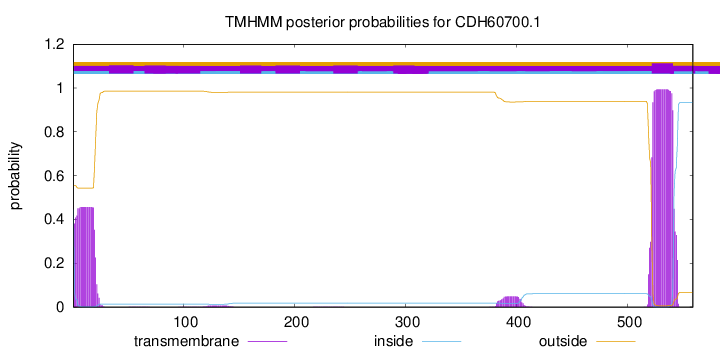
TMHMM annotations
Basic Information help
| Species | Lichtheimia corymbifera | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Mucoromycota; Mucoromycetes; ; Lichtheimiaceae; Lichtheimia; Lichtheimia corymbifera | |||||||||||
| CAZyme ID | CDH60700.1 | |||||||||||
| CAZy Family | GT69 | |||||||||||
| CAZyme Description | udp-glucosyl transferase family protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT1 | 264 | 461 | 2.3e-24 | 0.4293193717277487 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 340817 | GT1_Gtf-like | 7.87e-43 | 50 | 491 | 2 | 403 | UDP-glycosyltransferases and similar proteins. This family includes the Gtfs, a group of homologous glycosyltransferases involved in the final stages of the biosynthesis of antibiotics vancomycin and related chloroeremomycin. Gtfs transfer sugar moieties from an activated NDP-sugar donor to the oxidatively cross-linked heptapeptide core of vancomycin group antibiotics. The core structure is important for the bioactivity of the antibiotics. |
| 278624 | UDPGT | 4.25e-17 | 256 | 536 | 228 | 479 | UDP-glucoronosyl and UDP-glucosyl transferase. |
| 224732 | YjiC | 2.31e-16 | 51 | 493 | 4 | 399 | UDP:flavonoid glycosyltransferase YjiC, YdhE family [Carbohydrate transport and metabolism]. |
| 177813 | PLN02152 | 7.28e-16 | 290 | 479 | 248 | 438 | indole-3-acetate beta-glucosyltransferase |
| 223071 | egt | 1.21e-15 | 258 | 458 | 250 | 432 | ecdysteroid UDP-glucosyltransferase; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 6 | 559 | 5 | 551 | |
| 1.42e-239 | 32 | 550 | 23 | 541 | |
| 1.07e-179 | 37 | 530 | 30 | 522 | |
| 8.87e-129 | 185 | 534 | 11 | 353 | |
| 6.37e-128 | 185 | 543 | 17 | 369 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.48e-11 | 273 | 435 | 253 | 417 | Crystal structure of LpCGTa in complex with UDP [Landoltia punctata],6LG1_B Crystal structure of LpCGTa in complex with UDP [Landoltia punctata] |
|
| 5.78e-11 | 259 | 436 | 228 | 399 | Crystal Structure of Medicago truncatula UGT71G1 [Medicago truncatula],2ACV_B Crystal Structure of Medicago truncatula UGT71G1 [Medicago truncatula] |
|
| 5.81e-11 | 259 | 436 | 228 | 399 | Crystal Structure of Medicago truncatula UGT71G1 complexed with UDP-glucose [Medicago truncatula],2ACW_B Crystal Structure of Medicago truncatula UGT71G1 complexed with UDP-glucose [Medicago truncatula] |
|
| 1.06e-10 | 270 | 428 | 244 | 404 | Chain A, Glycosyltransferase [Calotropis gigantea],7W0K_A Chain A, Glycosyltransferase [Calotropis gigantea],7W0Z_A Chain A, Glycosyltransferase [Calotropis gigantea],7W10_A Chain A, Glycosyltransferase [Calotropis gigantea],7W11_A Chain A, Glycosyltransferase [Calotropis gigantea],7W1B_A Chain A, Glycosyltransferase [Calotropis gigantea],7W1H_A Chain A, Glycosyltransferase [Calotropis gigantea] |
|
| 2.30e-10 | 286 | 428 | 252 | 381 | Crystal structure of ugt transferase mutant in complex with UPG [Siraitia grosvenorii],6L90_A Crystal structure of ugt transferase enzyme [Siraitia grosvenorii] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.99e-13 | 258 | 427 | 225 | 391 | UDP-glycosyltransferase 71E1 OS=Stevia rebaudiana OX=55670 GN=UGT71E1 PE=2 SV=1 |
|
| 4.04e-13 | 255 | 523 | 250 | 499 | Putative UDP-glucuronosyltransferase ugt-50 OS=Caenorhabditis elegans OX=6239 GN=ugt-50 PE=1 SV=2 |
|
| 4.82e-13 | 264 | 431 | 236 | 408 | UDP-glycosyltransferase 71B2 OS=Arabidopsis thaliana OX=3702 GN=UGT71B2 PE=1 SV=1 |
|
| 1.80e-12 | 287 | 488 | 245 | 447 | UDP-glycosyltransferase 75B2 OS=Arabidopsis thaliana OX=3702 GN=UGT75B2 PE=2 SV=1 |
|
| 1.81e-12 | 301 | 487 | 272 | 444 | UDP-glycosyltransferase 74C1 OS=Arabidopsis thaliana OX=3702 GN=UGT74C1 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.001592 | 0.998378 | CS pos: 20-21. Pr: 0.9619 |