You are browsing environment: FUNGIDB
CAZyme Information: CDH60411.1
You are here: Home > Sequence: CDH60411.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
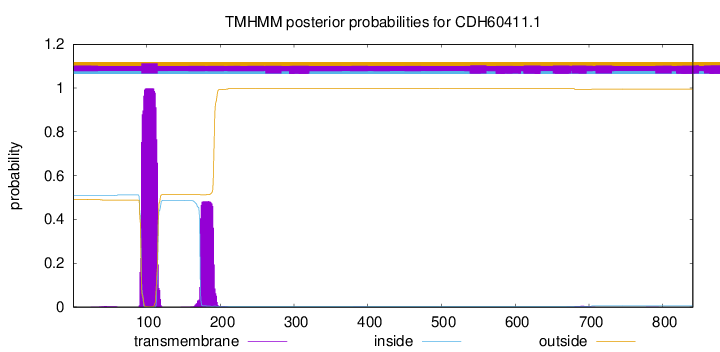
TMHMM annotations
Basic Information help
| Species | Lichtheimia corymbifera | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Mucoromycota; Mucoromycetes; ; Lichtheimiaceae; Lichtheimia; Lichtheimia corymbifera | |||||||||||
| CAZyme ID | CDH60411.1 | |||||||||||
| CAZy Family | GT59 | |||||||||||
| CAZyme Description | glycogen starch adp-glucose glycosyltransferasefamily 5 protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT5 | 306 | 770 | 2.1e-48 | 0.777542372881356 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 340822 | GT5_Glycogen_synthase_DULL1-like | 4.45e-32 | 166 | 808 | 2 | 473 | Glycogen synthase GlgA and similar proteins. This family is most closely related to the GT5 family of glycosyltransferases. Glycogen synthase (EC:2.4.1.21) catalyzes the formation and elongation of the alpha-1,4-glucose backbone using ADP-glucose, the second and key step of glycogen biosynthesis. This family includes starch synthases of plants, such as DULL1 in Zea mays and glycogen synthases of various organisms. |
| 223374 | GlgA | 2.49e-23 | 166 | 812 | 3 | 480 | Glycogen synthase [Carbohydrate transport and metabolism]. |
| 234809 | glgA | 2.33e-20 | 166 | 812 | 3 | 465 | glycogen synthase GlgA. |
| 404563 | Glyco_trans_1_4 | 3.85e-18 | 592 | 710 | 1 | 115 | Glycosyl transferases group 1. |
| 223515 | RfaB | 3.00e-17 | 414 | 814 | 20 | 381 | Glycosyltransferase involved in cell wall bisynthesis [Cell wall/membrane/envelope biogenesis]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 839 | 1 | 830 | |
| 9.45e-234 | 66 | 814 | 45 | 723 | |
| 4.57e-102 | 92 | 814 | 83 | 808 | |
| 3.63e-17 | 303 | 811 | 272 | 715 | |
| 6.98e-17 | 349 | 726 | 175 | 496 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.49e-11 | 537 | 724 | 161 | 379 | Sucrose-phosphate synthase (tll1590) from Thermosynechococcus elongatus [Thermosynechococcus vestitus],6KIH_B Sucrose-phosphate synthase (tll1590) from Thermosynechococcus elongatus [Thermosynechococcus vestitus],6KIH_C Sucrose-phosphate synthase (tll1590) from Thermosynechococcus elongatus [Thermosynechococcus vestitus],6KIH_D Sucrose-phosphate synthase (tll1590) from Thermosynechococcus elongatus [Thermosynechococcus vestitus],6KIH_E Sucrose-phosphate synthase (tll1590) from Thermosynechococcus elongatus [Thermosynechococcus vestitus],6KIH_F Sucrose-phosphate synthase (tll1590) from Thermosynechococcus elongatus [Thermosynechococcus vestitus],6KIH_G Sucrose-phosphate synthase (tll1590) from Thermosynechococcus elongatus [Thermosynechococcus vestitus],6KIH_H Sucrose-phosphate synthase (tll1590) from Thermosynechococcus elongatus [Thermosynechococcus vestitus],6KIH_I Sucrose-phosphate synthase (tll1590) from Thermosynechococcus elongatus [Thermosynechococcus vestitus],6KIH_J Sucrose-phosphate synthase (tll1590) from Thermosynechococcus elongatus [Thermosynechococcus vestitus],6KIH_K Sucrose-phosphate synthase (tll1590) from Thermosynechococcus elongatus [Thermosynechococcus vestitus],6KIH_L Sucrose-phosphate synthase (tll1590) from Thermosynechococcus elongatus [Thermosynechococcus vestitus] |
|
| 1.17e-10 | 564 | 707 | 12 | 154 | Structure of the C domain of glycogen synthase from Pyrococcus abyssi [Pyrococcus abyssi] |
|
| 6.43e-10 | 554 | 707 | 212 | 359 | XFEL crystal structure of human melatonin receptor MT1 in complex with ramelteon [Homo sapiens] |
|
| 6.43e-10 | 554 | 707 | 212 | 359 | XFEL crystal structure of human melatonin receptor MT1 in complex with 2-phenylmelatonin [Homo sapiens],6ME4_A XFEL crystal structure of human melatonin receptor MT1 in complex with 2-iodomelatonin [Homo sapiens],6ME5_A XFEL crystal structure of human melatonin receptor MT1 in complex with agomelatine [Homo sapiens],6PS8_A XFEL MT1R structure by ligand exchange from agomelatine to 2-phenylmelatonin. [Homo sapiens] |
|
| 6.64e-10 | 554 | 707 | 229 | 376 | Crystal structure of the Neurokinin 1 receptor in complex with the small molecule antagonist Aprepitant [Homo sapiens],6HLP_A Crystal structure of the Neurokinin 1 receptor in complex with the small molecule antagonist Netupitant [Homo sapiens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.22e-16 | 349 | 763 | 114 | 459 | Glycogen synthase OS=Protochlamydia amoebophila (strain UWE25) OX=264201 GN=glgA PE=3 SV=1 |
|
| 1.38e-14 | 446 | 709 | 189 | 405 | Glycogen synthase OS=Sulfurihydrogenibium sp. (strain YO3AOP1) OX=436114 GN=glgA PE=3 SV=1 |
|
| 1.03e-13 | 349 | 726 | 111 | 432 | Glycogen synthase 2 OS=Methylococcus capsulatus (strain ATCC 33009 / NCIMB 11132 / Bath) OX=243233 GN=glgA2 PE=3 SV=2 |
|
| 1.07e-13 | 382 | 710 | 146 | 427 | Glycogen synthase OS=Thermodesulfovibrio yellowstonii (strain ATCC 51303 / DSM 11347 / YP87) OX=289376 GN=glgA PE=3 SV=1 |
|
| 3.54e-13 | 451 | 708 | 160 | 412 | Probable glycogen synthase OS=Methanocaldococcus jannaschii (strain ATCC 43067 / DSM 2661 / JAL-1 / JCM 10045 / NBRC 100440) OX=243232 GN=glgA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
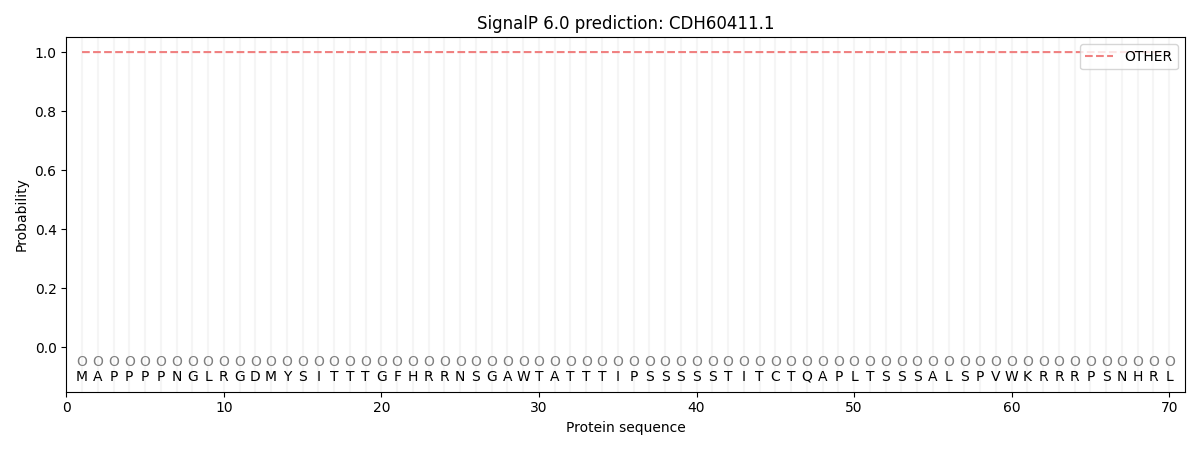
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000071 | 0.000000 |