You are browsing environment: FUNGIDB
CAZyme Information: CDH58997.1
You are here: Home > Sequence: CDH58997.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Lichtheimia corymbifera | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Mucoromycota; Mucoromycetes; ; Lichtheimiaceae; Lichtheimia; Lichtheimia corymbifera | |||||||||||
| CAZyme ID | CDH58997.1 | |||||||||||
| CAZy Family | GT4 | |||||||||||
| CAZyme Description | glycosyltransferase family 31 and 32 protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 2.4.1.149:9 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT31 | 286 | 471 | 2.9e-33 | 0.9739583333333334 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 250845 | Galactosyl_T | 2.55e-22 | 287 | 472 | 3 | 193 | Galactosyltransferase. This family includes the galactosyltransferases UDP-galactose:2-acetamido-2-deoxy-D-glucose3beta-galactosyltransferase and UDP-Gal:beta-GlcNAc beta 1,3-galactosyltranferase. Specific galactosyltransferases transfer galactose to GlcNAc terminal chains in the synthesis of the lacto-series oligosaccharides types 1 and 2. |
| 403517 | EOS1 | 1.54e-12 | 112 | 201 | 3 | 94 | N-glycosylation protein. This family is not required for survival of S.cerevisiae, but its deletion leads to heightened sensitivity to oxidative stress. It appears to be involved in N-glycosylation, and resides in the endoplasmic reticulum. |
| 178735 | PLN03193 | 2.09e-09 | 310 | 477 | 181 | 353 | beta-1,3-galactosyltransferase; Provisional |
| 215596 | PLN03133 | 1.66e-08 | 231 | 492 | 349 | 611 | beta-1,3-galactosyltransferase; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 40 | 565 | 1 | 522 | |
| 2.04e-144 | 44 | 559 | 4 | 510 | |
| 3.13e-106 | 26 | 562 | 6 | 585 | |
| 5.37e-19 | 271 | 500 | 57 | 286 | |
| 1.49e-17 | 255 | 491 | 24 | 257 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.41e-15 | 262 | 476 | 48 | 261 | Beta-1,3-galactosyltransferase 6 OS=Homo sapiens OX=9606 GN=B3GALT6 PE=1 SV=2 |
|
| 3.82e-15 | 240 | 441 | 50 | 263 | Lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase OS=Sus scrofa OX=9823 GN=B3GNT5 PE=2 SV=1 |
|
| 9.83e-15 | 260 | 492 | 362 | 596 | Hydroxyproline O-galactosyltransferase GALT3 OS=Arabidopsis thaliana OX=3702 GN=GALT3 PE=2 SV=1 |
|
| 5.44e-14 | 329 | 490 | 312 | 475 | UDP-GalNAc:beta-1,3-N-acetylgalactosaminyltransferase 2 OS=Mus musculus OX=10090 GN=B3galnt2 PE=1 SV=1 |
|
| 5.89e-14 | 270 | 476 | 50 | 257 | Beta-1,3-galactosyltransferase 6 OS=Mus musculus OX=10090 GN=B3galt6 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000038 | 0.000002 |
TMHMM Annotations download full data without filtering help
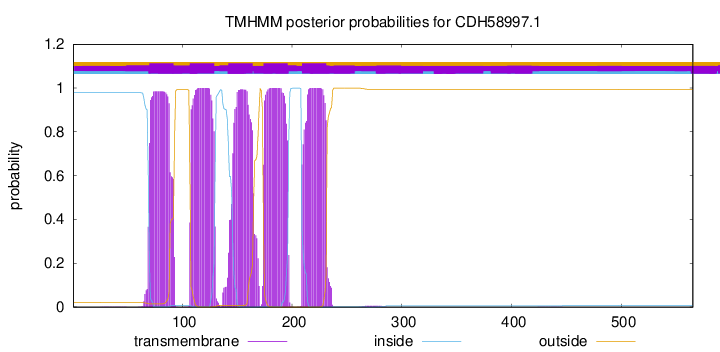
| Start | End |
|---|---|
| 70 | 92 |
| 107 | 129 |
| 142 | 164 |
| 174 | 196 |
| 209 | 231 |
