You are browsing environment: FUNGIDB
CAZyme Information: CDH58828.1
You are here: Home > Sequence: CDH58828.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
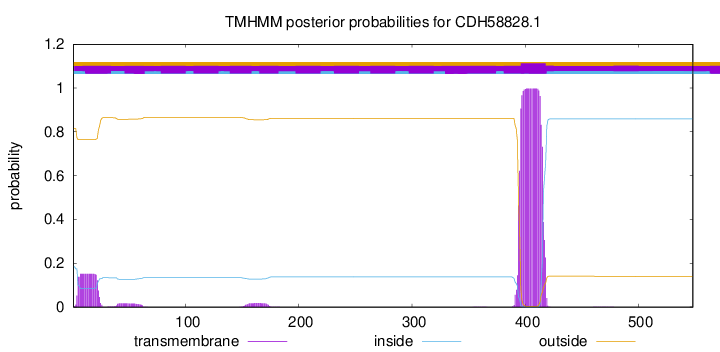
TMHMM annotations
Basic Information help
| Species | Lichtheimia corymbifera | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Mucoromycota; Mucoromycetes; ; Lichtheimiaceae; Lichtheimia; Lichtheimia corymbifera | |||||||||||
| CAZyme ID | CDH58828.1 | |||||||||||
| CAZy Family | GT39 | |||||||||||
| CAZyme Description | lysophospholipase a | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE16 | 47 | 287 | 8.7e-45 | 0.8838951310861424 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 238882 | fatty_acyltransferase_like | 2.12e-31 | 46 | 284 | 3 | 230 | Fatty acyltransferase-like subfamily of the SGNH hydrolases, a diverse family of lipases and esterases. The tertiary fold of the enzyme is substantially different from that of the alpha/beta hydrolase family and unique among all known hydrolases; its active site closely resembles the Ser-His-Asp(Glu) triad found in other serine hydrolases. Might catalyze fatty acid transfer between phosphatidylcholine and sterols. |
| 409004 | DPBB_RlpA_EXP_N-like | 1.36e-20 | 458 | 531 | 3 | 70 | double-psi beta-barrel fold of RlpA, N-terminal domain of expansins, and similar domains. The double-psi beta-barrel (DPBB) fold is found in a divergent group of proteins, including endolytic peptidoglycan transglycosylase RlpA (rare lipoprotein A), EG45-like domain containing proteins, kiwellins, Streptomyces papain inhibitor (SPI), and the N-terminal domain of plant and bacterial expansins. RlpA may work in tandem with amidases to degrade peptidoglycan (PG) in the division septum and lateral wall to facilitate daughter cell separation. An EG45-like domain containing protein from Arabidopsis thaliana, called plant natriuretic peptide A (AtPNP-A), functions in cell volume regulation. Kiwellin proteins comprise a widespread family of plant-defense proteins that target pathogenic bacterial/fungal effectors that down-regulate plant defense responses. SPI is a stress protein produced under hyperthermal stress conditions that serves as a glutamine and lysine donor substrate for microbial transglutaminase (MTG, EC 2.3.2.13) from Streptomycetes. Some expansin family proteins display cell wall loosening activity and are involved in cell expansion and other developmental events during which cell wall modification occurs. |
| 238875 | SGNH_plant_lipase_like | 1.58e-11 | 113 | 282 | 79 | 265 | SGNH_plant_lipase_like, a plant specific subfamily of the SGNH-family of hydrolases, a diverse family of lipases and esterases. The tertiary fold of the enzyme is substantially different from that of the alpha/beta hydrolase family and unique among all known hydrolases; its active site closely resembles the Ser-His-Asp(Glu) triad found in other serine hydrolases. |
| 409010 | DPBB_SPI-like | 1.76e-09 | 456 | 527 | 1 | 72 | double-psi beta-barrel fold of Streptomyces papain inhibitor and similar proteins. Streptomyces papain inhibitor (SPI) adopts a rigid, thermo-resistant double-psi-beta-barrel (DPBB) fold that is stabilized by two cysteine bridges. SPI serves as a glutamine and lysine donor substrate for microbial transglutaminase (MTG, EC 2.3.2.13) from Streptomycetes, that is used to covalently and specifically link functional amines to glutamine donor sites of therapeutic proteins. SPI is a stress protein produced under hyperthermal stress conditions, and is able to inhibit the cysteine proteases, papain and bromelain, as well as the bovine serine protease trypsin. |
| 395531 | Lipase_GDSL | 1.99e-06 | 46 | 272 | 2 | 202 | GDSL-like Lipase/Acylhydrolase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.87e-211 | 1 | 304 | 1 | 300 | |
| 2.89e-44 | 10 | 302 | 6 | 309 | |
| 5.52e-43 | 13 | 302 | 10 | 308 | |
| 1.71e-40 | 30 | 313 | 45 | 334 | |
| 3.88e-40 | 15 | 169 | 19 | 165 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
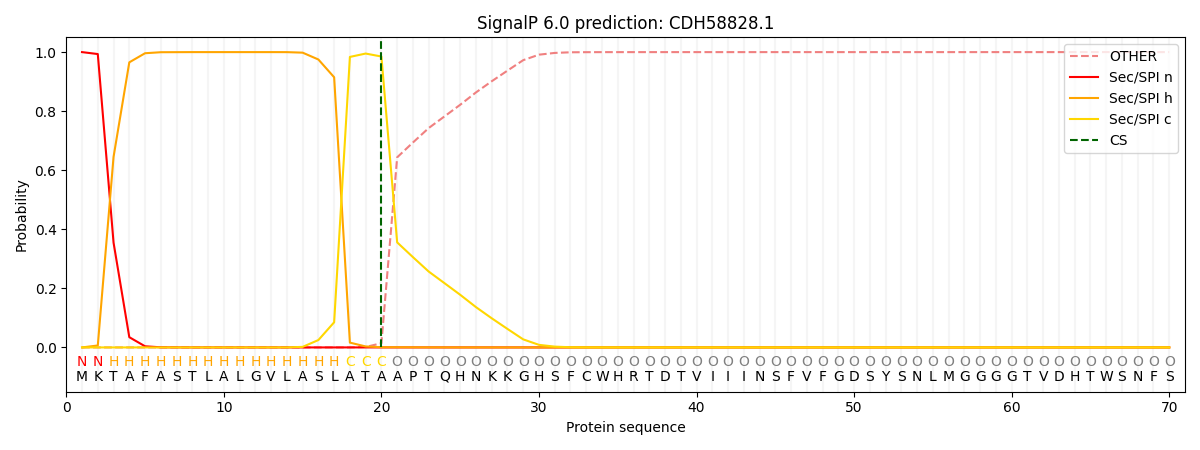
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000201 | 0.999768 | CS pos: 20-21. Pr: 0.9850 |