You are browsing environment: FUNGIDB
CAZyme Information: CDH58441.1
You are here: Home > Sequence: CDH58441.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
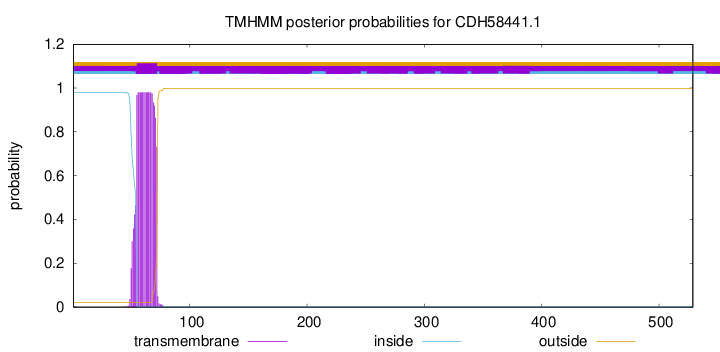
TMHMM annotations
Basic Information help
| Species | Lichtheimia corymbifera | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Mucoromycota; Mucoromycetes; ; Lichtheimiaceae; Lichtheimia; Lichtheimia corymbifera | |||||||||||
| CAZyme ID | CDH58441.1 | |||||||||||
| CAZy Family | GT3 | |||||||||||
| CAZyme Description | glycosyltransferase-like protein large1 | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 2.4.1.-:23 | 2.4.2.-:23 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT49 | 211 | 502 | 2.6e-43 | 0.8367952522255193 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 404735 | Glyco_transf_49 | 1.97e-32 | 216 | 499 | 1 | 326 | Glycosyl-transferase for dystroglycan. This glycosyl-transferase brings about the glycosylation of the alpha-dystroglycan subunit. Dystroglycan is an integral member of the skeletal muscular dystrophin glycoprotein complex, which links dystrophin to proteins in the extracellular matrix. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 529 | 1 | 529 | |
| 2.67e-65 | 115 | 514 | 118 | 557 | |
| 4.96e-61 | 120 | 514 | 120 | 567 | |
| 9.10e-32 | 195 | 465 | 145 | 400 | |
| 4.03e-27 | 216 | 501 | 341 | 613 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 8.73e-28 | 216 | 501 | 402 | 674 | Xylosyl- and glucuronyltransferase LARGE2 OS=Mus musculus OX=10090 GN=Large2 PE=1 SV=1 |
|
| 9.12e-26 | 216 | 501 | 401 | 674 | Xylosyl- and glucuronyltransferase LARGE2 OS=Rattus norvegicus OX=10116 GN=Large2 PE=2 SV=1 |
|
| 2.44e-25 | 195 | 500 | 454 | 744 | Xylosyl- and glucuronyltransferase LARGE1 OS=Danio rerio OX=7955 GN=large1 PE=2 SV=1 |
|
| 4.11e-25 | 216 | 511 | 431 | 705 | Xylosyl- and glucuronyltransferase LARGE2 OS=Homo sapiens OX=9606 GN=LARGE2 PE=1 SV=2 |
|
| 3.17e-24 | 216 | 501 | 456 | 727 | Xylosyl- and glucuronyltransferase LARGE2s OS=Gallus gallus OX=9031 GN=LARGE2 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
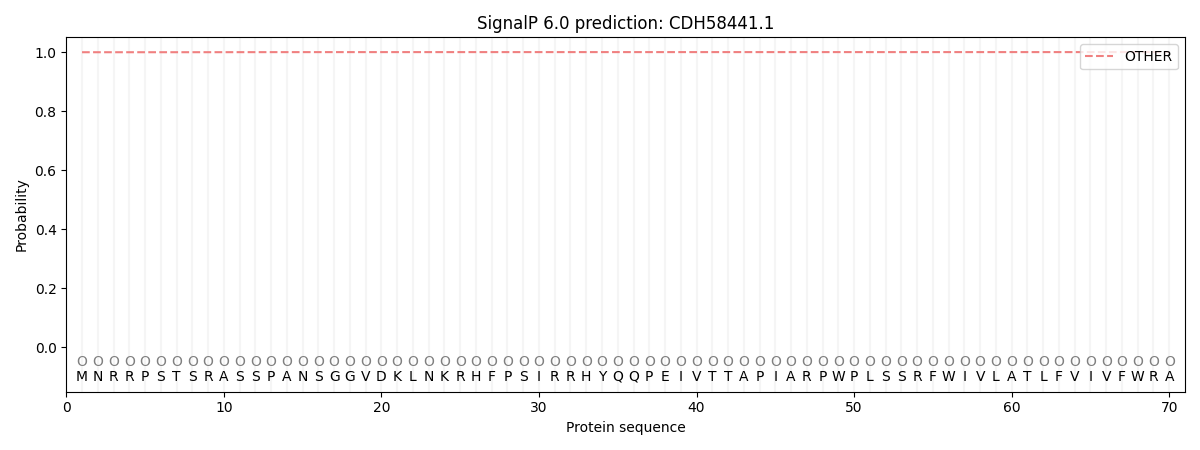
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.999725 | 0.000277 |