You are browsing environment: FUNGIDB
CAZyme Information: CDH58423.1
You are here: Home > Sequence: CDH58423.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Lichtheimia corymbifera | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Mucoromycota; Mucoromycetes; ; Lichtheimiaceae; Lichtheimia; Lichtheimia corymbifera | |||||||||||
| CAZyme ID | CDH58423.1 | |||||||||||
| CAZy Family | GT3 | |||||||||||
| CAZyme Description | l-ascorbate oxidase-like | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA1 | 50 | 563 | 1.2e-92 | 0.9692737430167597 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 132431 | ascorbOXfungal | 2.01e-75 | 31 | 569 | 11 | 531 | L-ascorbate oxidase, fungal type. This model describes a family of fungal ascorbate oxidases, within a larger family of multicopper oxidases that also includes plant ascorbate oxidases (TIGR03388), plant laccases and laccase-like proteins (TIGR03389), and related proteins. The member from Acremonium sp. HI-25 is characterized. |
| 274555 | ascorbase | 3.01e-62 | 28 | 563 | 1 | 517 | L-ascorbate oxidase, plant type. Members of this protein family are the copper-containing enzyme L-ascorbate oxidase (EC 1.10.3.3), also called ascorbase. This family is found in flowering plants, and shows greater sequence similarity to a family of laccases (EC 1.10.3.2) from plants than to other known ascorbate oxidases. |
| 177843 | PLN02191 | 1.10e-58 | 42 | 574 | 37 | 551 | L-ascorbate oxidase |
| 215324 | PLN02604 | 1.11e-57 | 26 | 563 | 22 | 540 | oxidoreductase |
| 274556 | laccase | 4.38e-52 | 26 | 569 | 1 | 519 | laccase, plant. Members of this protein family include the copper-containing enzyme laccase (EC 1.10.3.2), often several from a single plant species, and additional, uncharacterized, closely related plant proteins termed laccase-like multicopper oxidases. This protein family shows considerable sequence similarity to the L-ascorbate oxidase (EC 1.10.3.3) family. Laccases are enzymes of rather broad specificity, and classification of all proteins scoring about the trusted cutoff of this model as laccases may be appropriate. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 575 | 1 | 574 | |
| 3.40e-151 | 1 | 540 | 1 | 518 | |
| 4.72e-136 | 31 | 574 | 18 | 570 | |
| 1.33e-133 | 21 | 574 | 18 | 645 | |
| 3.69e-59 | 33 | 575 | 35 | 521 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.95e-44 | 42 | 574 | 17 | 528 | Refined Crystal Structure Of Ascorbate Oxidase At 1.9 Angstroms Resolution [Cucurbita pepo var. melopepo],1AOZ_B Refined Crystal Structure Of Ascorbate Oxidase At 1.9 Angstroms Resolution [Cucurbita pepo var. melopepo],1ASO_A X-Ray Structures And Mechanistic Implications Of Three Functional Derivatives Of Ascorbate Oxidase From Zucchini: Reduced-, Peroxide-, And Azide-Forms [Cucurbita pepo var. melopepo],1ASO_B X-Ray Structures And Mechanistic Implications Of Three Functional Derivatives Of Ascorbate Oxidase From Zucchini: Reduced-, Peroxide-, And Azide-Forms [Cucurbita pepo var. melopepo],1ASP_A X-ray Structures And Mechanistic Implications Of Three Functional Derivatives Of Ascorbate Oxidase From Zucchini: Reduced-, Peroxide-, And Azide-forms [Cucurbita pepo var. melopepo],1ASP_B X-ray Structures And Mechanistic Implications Of Three Functional Derivatives Of Ascorbate Oxidase From Zucchini: Reduced-, Peroxide-, And Azide-forms [Cucurbita pepo var. melopepo],1ASQ_A X-Ray Structures And Mechanistic Implications Of Three Functional Derivatives Of Ascorbate Oxidase From Zucchini: Reduced-, Peroxide-, And Azide-Forms [Cucurbita pepo var. melopepo],1ASQ_B X-Ray Structures And Mechanistic Implications Of Three Functional Derivatives Of Ascorbate Oxidase From Zucchini: Reduced-, Peroxide-, And Azide-Forms [Cucurbita pepo var. melopepo] |
|
| 8.94e-41 | 33 | 572 | 9 | 469 | Chain A, LACCASE 1 [Coprinopsis cinerea] |
|
| 9.07e-41 | 33 | 572 | 9 | 469 | Chain A, Laccase [Coprinopsis cinerea] |
|
| 1.92e-40 | 47 | 571 | 20 | 481 | Crystal Structure of Fet3p, a Multicopper Oxidase that Functions in Iron Import [Saccharomyces cerevisiae],1ZPU_B Crystal Structure of Fet3p, a Multicopper Oxidase that Functions in Iron Import [Saccharomyces cerevisiae],1ZPU_C Crystal Structure of Fet3p, a Multicopper Oxidase that Functions in Iron Import [Saccharomyces cerevisiae],1ZPU_D Crystal Structure of Fet3p, a Multicopper Oxidase that Functions in Iron Import [Saccharomyces cerevisiae],1ZPU_E Crystal Structure of Fet3p, a Multicopper Oxidase that Functions in Iron Import [Saccharomyces cerevisiae],1ZPU_F Crystal Structure of Fet3p, a Multicopper Oxidase that Functions in Iron Import [Saccharomyces cerevisiae] |
|
| 5.38e-40 | 32 | 572 | 28 | 489 | Crystal structure of laccases from Pycnoporus sanguineus, izoform I [Trametes sanguinea] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.20e-49 | 26 | 563 | 28 | 544 | L-ascorbate oxidase OS=Nicotiana tabacum OX=4097 GN=AAO PE=2 SV=1 |
|
| 7.78e-49 | 87 | 565 | 1 | 484 | Multicopper oxidase terE OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=terE PE=1 SV=1 |
|
| 3.12e-48 | 33 | 573 | 35 | 597 | Laccase-like multicopper oxidase 1 OS=Myceliophthora thermophila (strain ATCC 42464 / BCRC 31852 / DSM 1799) OX=573729 GN=LMCO1 PE=1 SV=1 |
|
| 3.53e-47 | 42 | 574 | 37 | 548 | L-ascorbate oxidase OS=Arabidopsis thaliana OX=3702 GN=AAO PE=1 SV=1 |
|
| 6.62e-46 | 28 | 574 | 61 | 565 | Laccase-1 OS=Cryptococcus neoformans var. neoformans serotype D (strain B-3501A) OX=283643 GN=LAC1 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
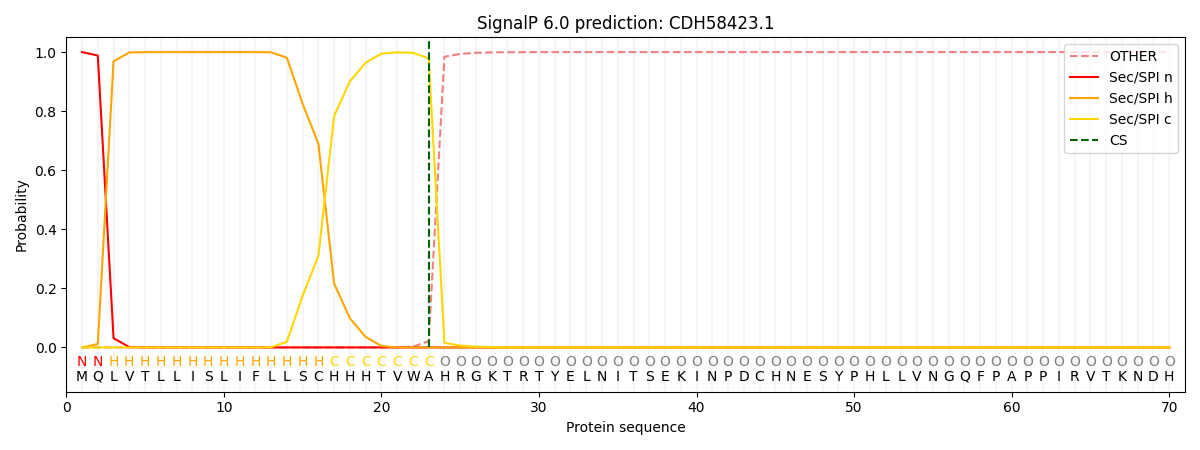
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000182 | 0.999793 | CS pos: 23-24. Pr: 0.9791 |
