You are browsing environment: FUNGIDB
CAZyme Information: CDH56583.1
You are here: Home > Sequence: CDH56583.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Lichtheimia corymbifera | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Mucoromycota; Mucoromycetes; ; Lichtheimiaceae; Lichtheimia; Lichtheimia corymbifera | |||||||||||
| CAZyme ID | CDH56583.1 | |||||||||||
| CAZy Family | GT2 | |||||||||||
| CAZyme Description | aldo keto reductase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH46 | 86 | 301 | 4.4e-41 | 0.9504504504504504 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 381369 | AKR_AKR6C1_2 | 1.67e-173 | 373 | 696 | 1 | 319 | AKR6C family of aldo-keto reductase (AKR). Voltage-gated potassium channel subunit beta (KCAB) from Arabidopsis thaliana and Egeria densa are founding members of aldo-keto reductase family 6 member C1 (AKR6C1) and C2 (AKR6C2), respectively. KCAB, also called Shaker channel b-subunit, or K(+) channel subunit beta, or potassium voltage beta 1, or KV-beta1, or KAB1, is a probable accessory potassium channel protein which modulates the activity of the pore-forming alpha subunit. |
| 381300 | Aldo_ket_red_shaker-like | 2.41e-139 | 382 | 682 | 1 | 292 | Shaker potassium channel beta subunit family and similar proteins. This family includes voltage-gated potassium channel subunits, beta-1 (KCAB1B), beta-2 (KCAB2B) and beta-3 (KCAB3B). KCAB1B and KCAB2B are cytoplasmic potassium channel subunits that modulate the characteristics of the channel-forming alpha-subunits. KCAB3B is an accessory potassium channel protein which modulates the activity of the pore-forming alpha subunit. The family also includes Drosophila melanogaster Hk protein, a founding member of aldo-keto reductase family 6 member B1 (AKR6B1), as well as voltage-gated potassium channel subunit beta (KCAB) from Arabidopsis thaliana and Egeria densa, founding members of AKR6C1and AKR6C2, respectively. Hk protein, also called hyperkinetic, is a beta subunit of Shaker (Sh) K+ channels and shows high sequence homology to aldoketoreductase. KCAB, also called Shaker channel b-subunit, or K(+) channel subunit beta, or potassium voltage beta 1, or KV-beta1, or KAB1, is a probable accessory potassium channel protein which modulates the activity of the pore-forming alpha subunit. |
| 381367 | Aldo_ket_red_shaker | 2.14e-129 | 375 | 689 | 2 | 310 | Shaker potassium channel beta subunit (AKR6A) family of aldo-keto reductase (AKR). This family includes voltage-gated potassium channel subunits, beta-1 (KCAB1B), beta-2 (KCAB2B) and beta-3 (KCAB3B). KCAB1B and KCAB2B are cytoplasmic potassium channel subunits that modulate the characteristics of the channel-forming alpha-subunits. KCAB3B is an accessory potassium channel protein which modulates the activity of the pore-forming alpha subunit. |
| 381385 | AKR_KCAB1B_AKR6A3-like | 8.80e-119 | 373 | 701 | 1 | 323 | voltage-gated potassium channel subunit beta-1 (KCAB1B) and similar proteins. KCAB1B from Homo sapiens, Mus musculus, Mustela putorius, Rattus norvegicus, and Kvb1.1, Kvb1.2 from Oryctolagus cuniculus, are founding members of aldo-keto reductase family 6 member A3 (AKR6A3), A8 (AKR6A8), A10a (AKR6A10a), A13 (AKR6A13), A7 (AKR6A7) and A10b (AKR6A10b), respectively. KCAB1B, also called Shaker channel b-subunit 1(Kvb1), K(+) channel subunit beta-1, or Kv-beta-1, is a cytoplasmic potassium channel subunit that modulates the characteristics of the channel-forming alpha-subunits. It modulates action potentials via its effect on the pore-forming alpha subunits. |
| 381386 | AKR_KCAB3B_AKR6A9-like | 2.01e-116 | 373 | 701 | 3 | 325 | voltage-gated potassium channel subunit beta-3 (KCAB3B) and similar proteins. KCAB3B from Homo sapiens, Rattus norvegicus, and Mus musculus, are founding members of aldo-keto reductase family 6 member A9 (AKR6A9), A12 (AKR6A12), A14 (AKR6A14), respectively. KCAB3B, also called Shaker channel b-subunit 3 (Kvb3), K(+) channel subunit beta-3, or Kv-beta-3, is an accessory potassium channel protein which modulates the activity of the pore-forming alpha subunit. It alters the functional properties of Kv1.5. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 9.49e-242 | 1 | 361 | 1 | 365 | |
| 1.85e-64 | 67 | 334 | 58 | 325 | |
| 1.94e-46 | 373 | 704 | 55 | 378 | |
| 1.54e-28 | 89 | 318 | 9 | 237 | |
| 3.62e-28 | 89 | 318 | 45 | 273 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 9.31e-107 | 375 | 708 | 4 | 331 | Chain A, KV BETA2 PROTEIN [Rattus norvegicus] |
|
| 9.63e-107 | 375 | 708 | 5 | 332 | Mammalian Shaker Kv1.2 potassium channel- beta subunit complex [Rattus norvegicus],2R9R_A Shaker family voltage dependent potassium channel (kv1.2-kv2.1 paddle chimera channel) in association with beta subunit [Rattus norvegicus],2R9R_G Shaker family voltage dependent potassium channel (kv1.2-kv2.1 paddle chimera channel) in association with beta subunit [Rattus norvegicus],3LNM_A Chain A, Voltage-gated potassium channel subunit beta-2 [Rattus norvegicus],3LNM_C Chain C, Voltage-gated potassium channel subunit beta-2 [Rattus norvegicus],4JTA_A Crystal structure of Kv1.2-2.1 paddle chimera channel in complex with Charybdotoxin [Rattus norvegicus],4JTA_P Crystal structure of Kv1.2-2.1 paddle chimera channel in complex with Charybdotoxin [Rattus norvegicus],4JTC_A Crystal structure of Kv1.2-2.1 paddle chimera channel in complex with Charybdotoxin in Cs+ [Rattus norvegicus],4JTC_G Crystal structure of Kv1.2-2.1 paddle chimera channel in complex with Charybdotoxin in Cs+ [Rattus norvegicus],4JTD_A Crystal structure of Kv1.2-2.1 paddle chimera channel in complex with Lys27Met mutant of Charybdotoxin [Rattus norvegicus],4JTD_G Crystal structure of Kv1.2-2.1 paddle chimera channel in complex with Lys27Met mutant of Charybdotoxin [Rattus norvegicus],5WIE_A Crystal structure of a Kv1.2-2.1 chimera K+ channel V406W mutant in an inactivated state [Rattus norvegicus],5WIE_G Crystal structure of a Kv1.2-2.1 chimera K+ channel V406W mutant in an inactivated state [Rattus norvegicus],7SIT_A Chain A, Voltage-gated potassium channel subunit beta-2 [Rattus norvegicus],7SIT_C Chain C, Voltage-gated potassium channel subunit beta-2 [Rattus norvegicus],7SIZ_A Chain A, Voltage-gated potassium channel subunit beta-2 [Rattus norvegicus],7SIZ_C Chain C, Voltage-gated potassium channel subunit beta-2 [Rattus norvegicus] |
|
| 9.63e-107 | 375 | 708 | 5 | 332 | The voltage-activated Kv1.2-2.1 paddle chimera channel in lipid nanodiscs [Rattus norvegicus],6EBK_C The voltage-activated Kv1.2-2.1 paddle chimera channel in lipid nanodiscs [Rattus norvegicus],6EBK_E The voltage-activated Kv1.2-2.1 paddle chimera channel in lipid nanodiscs [Rattus norvegicus],6EBK_G The voltage-activated Kv1.2-2.1 paddle chimera channel in lipid nanodiscs [Rattus norvegicus],6EBL_A The voltage-activated Kv1.2-2.1 paddle chimera channel in lipid nanodiscs, cytosolic domain [Rattus norvegicus],6EBL_C The voltage-activated Kv1.2-2.1 paddle chimera channel in lipid nanodiscs, cytosolic domain [Rattus norvegicus],6EBL_E The voltage-activated Kv1.2-2.1 paddle chimera channel in lipid nanodiscs, cytosolic domain [Rattus norvegicus],6EBL_G The voltage-activated Kv1.2-2.1 paddle chimera channel in lipid nanodiscs, cytosolic domain [Rattus norvegicus] |
|
| 2.09e-106 | 375 | 708 | 39 | 366 | Chain A, Voltage-gated potassium channel subunit beta-2 [Homo sapiens],7EJ1_C Chain C, Voltage-gated potassium channel subunit beta-2 [Homo sapiens],7EJ1_E Chain E, Voltage-gated potassium channel subunit beta-2 [Homo sapiens],7EJ1_G Chain G, Voltage-gated potassium channel subunit beta-2 [Homo sapiens],7EJ2_A Chain A, Voltage-gated potassium channel subunit beta-2 [Homo sapiens],7EJ2_C Chain C, Voltage-gated potassium channel subunit beta-2 [Homo sapiens],7EJ2_E Chain E, Voltage-gated potassium channel subunit beta-2 [Homo sapiens],7EJ2_G Chain G, Voltage-gated potassium channel subunit beta-2 [Homo sapiens] |
|
| 2.95e-106 | 375 | 708 | 39 | 366 | Chain A, Voltage-gated potassium channel subunit beta-2 [Rattus norvegicus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.38e-106 | 375 | 708 | 39 | 366 | Voltage-gated potassium channel subunit beta-2 OS=Xenopus laevis OX=8355 GN=kcnab2 PE=2 SV=1 |
|
| 1.38e-106 | 375 | 708 | 39 | 366 | Voltage-gated potassium channel subunit beta-2 OS=Bos taurus OX=9913 GN=KCNAB2 PE=1 SV=1 |
|
| 1.08e-105 | 375 | 708 | 39 | 366 | Voltage-gated potassium channel subunit beta-2 OS=Homo sapiens OX=9606 GN=KCNAB2 PE=1 SV=2 |
|
| 1.16e-105 | 373 | 708 | 71 | 400 | Voltage-gated potassium channel subunit beta-1 OS=Gallus gallus OX=9031 GN=KCNAB1 PE=2 SV=1 |
|
| 1.52e-105 | 375 | 708 | 39 | 366 | Voltage-gated potassium channel subunit beta-2 OS=Rattus norvegicus OX=10116 GN=Kcnab2 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
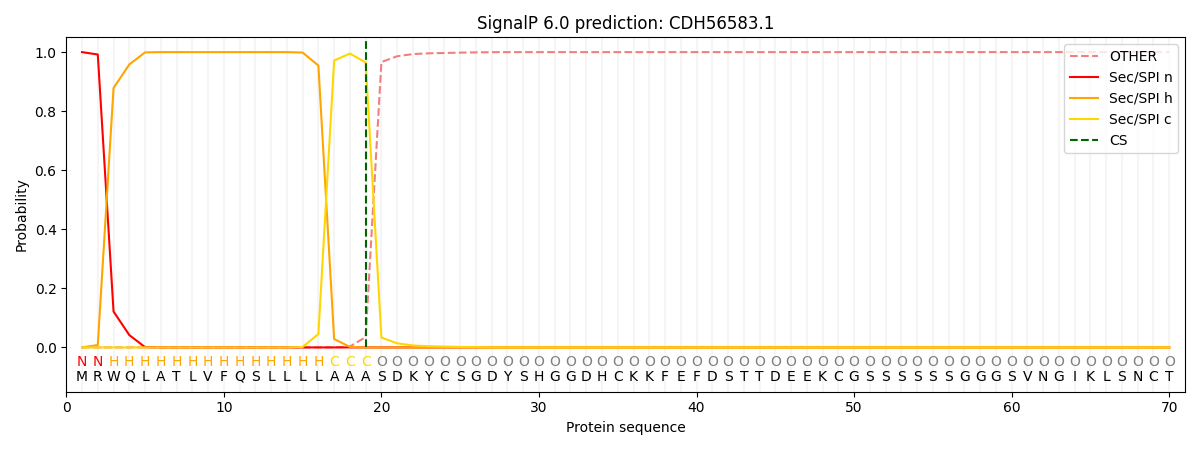
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000204 | 0.999758 | CS pos: 19-20. Pr: 0.9649 |
