You are browsing environment: FUNGIDB
CAZyme Information: CDH53161.1
You are here: Home > Sequence: CDH53161.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Lichtheimia corymbifera | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Mucoromycota; Mucoromycetes; ; Lichtheimiaceae; Lichtheimia; Lichtheimia corymbifera | |||||||||||
| CAZyme ID | CDH53161.1 | |||||||||||
| CAZy Family | GH28 | |||||||||||
| CAZyme Description | glycoside hydrolase family 28 protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.67:7 | 3.2.1.-:2 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH28 | 39 | 384 | 5.4e-77 | 0.963076923076923 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 395231 | Glyco_hydro_28 | 3.70e-54 | 39 | 371 | 1 | 310 | Glycosyl hydrolases family 28. Glycosyl hydrolase family 28 includes polygalacturonase EC:3.2.1.15 as well as rhamnogalacturonase A(RGase A), EC:3.2.1.-. These enzymes are important in cell wall metabolism. |
| 215426 | PLN02793 | 3.06e-35 | 10 | 367 | 43 | 397 | Probable polygalacturonase |
| 177865 | PLN02218 | 9.22e-34 | 27 | 364 | 82 | 408 | polygalacturonase ADPG |
| 215120 | PLN02188 | 1.55e-32 | 22 | 365 | 45 | 375 | polygalacturonase/glycoside hydrolase family protein |
| 178580 | PLN03003 | 4.86e-31 | 27 | 397 | 38 | 394 | Probable polygalacturonase At3g15720 |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 9.62e-291 | 2 | 397 | 9 | 403 | |
| 2.09e-215 | 1 | 394 | 8 | 398 | |
| 1.70e-77 | 22 | 396 | 70 | 447 | |
| 7.11e-77 | 22 | 393 | 81 | 455 | |
| 2.62e-73 | 17 | 395 | 51 | 436 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.54e-40 | 24 | 354 | 27 | 351 | Crystal structure of endo-xylogalacturonan hydrolase from Aspergillus tubingensis [Aspergillus tubingensis] |
|
| 3.36e-30 | 57 | 391 | 37 | 344 | Chain A, endo-polygalacturonase [Evansstolkia leycettana] |
|
| 3.36e-30 | 57 | 391 | 37 | 344 | Chain A, Endo-polygalacturonase [Evansstolkia leycettana] |
|
| 1.06e-29 | 57 | 391 | 29 | 336 | Chain A, Endo-polygalacturonase [Evansstolkia leycettana] |
|
| 5.49e-26 | 32 | 359 | 11 | 326 | Endopolygalacturonase from the phytopathogenic fungus Fusarium moniliforme [Fusarium verticillioides] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.84e-73 | 14 | 391 | 47 | 430 | Probable exopolygalacturonase B OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=pgxB PE=3 SV=1 |
|
| 9.61e-73 | 22 | 396 | 58 | 435 | Probable exopolygalacturonase B OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=pgxB PE=3 SV=1 |
|
| 1.29e-72 | 10 | 396 | 45 | 441 | Exopolygalacturonase OS=Cochliobolus carbonum OX=5017 GN=PGX1 PE=3 SV=1 |
|
| 1.35e-72 | 22 | 396 | 58 | 435 | Probable exopolygalacturonase B OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=pgxB PE=3 SV=2 |
|
| 6.06e-72 | 14 | 391 | 48 | 431 | Probable exopolygalacturonase B OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=pgxB PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
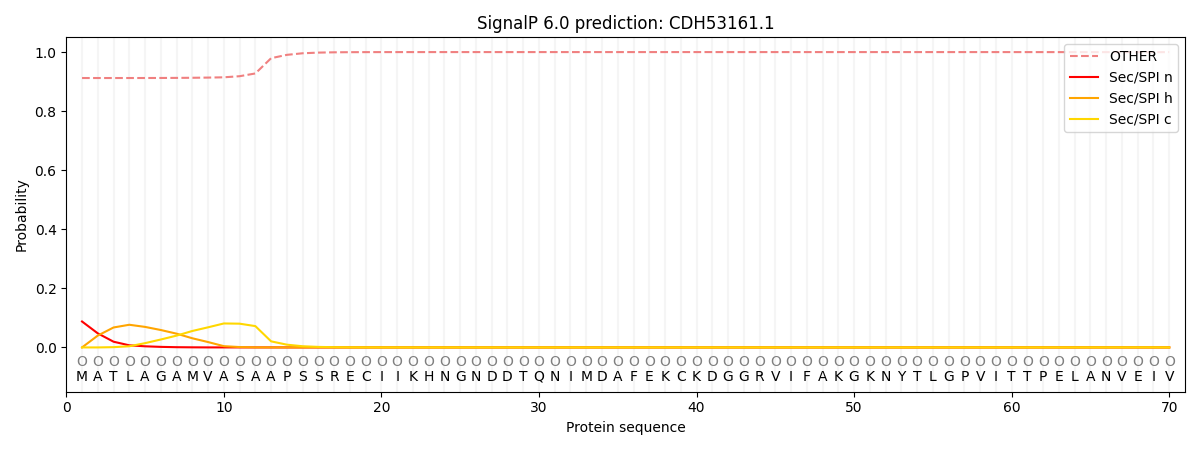
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.915988 | 0.084063 |
