You are browsing environment: FUNGIDB
CAZyme Information: CDH52275.1
You are here: Home > Sequence: CDH52275.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Lichtheimia corymbifera | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Mucoromycota; Mucoromycetes; ; Lichtheimiaceae; Lichtheimia; Lichtheimia corymbifera | |||||||||||
| CAZyme ID | CDH52275.1 | |||||||||||
| CAZy Family | GH18 | |||||||||||
| CAZyme Description | polysaccharide lyase family 14 protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 4.2.2.26:2 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL14 | 81 | 288 | 7.8e-68 | 0.9842105263157894 |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.49e-214 | 1 | 292 | 1 | 290 | |
| 2.58e-120 | 32 | 292 | 28 | 289 | |
| 9.60e-66 | 1 | 290 | 1 | 263 | |
| 3.96e-65 | 67 | 290 | 46 | 264 | |
| 3.96e-65 | 67 | 290 | 46 | 264 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.96e-49 | 67 | 290 | 43 | 272 | Crystal structure of the marine PL-14 alginate lyase from Aplysia kurodai [Aplysia kurodai],5GMT_B Crystal structure of the marine PL-14 alginate lyase from Aplysia kurodai [Aplysia kurodai] |
|
| 2.04e-15 | 82 | 289 | 39 | 241 | Crystal structure of D-glucuronic acid-bound alginate lyase vAL-1 from Chlorella virus [Paramecium bursaria Chlorella virus CVK2],3GNE_A Crystal structure of alginate lyase vAL-1 from Chlorella virus [Paramecium bursaria Chlorella virus CVK2],3GNE_B Crystal structure of alginate lyase vAL-1 from Chlorella virus [Paramecium bursaria Chlorella virus CVK2],3IM0_A Crystal structure of Chlorella virus vAL-1 soaked in 200mM D-glucuronic acid, 10% PEG-3350, and 200mM glycine-NaOH (pH 10.0) [Paramecium bursaria Chlorella virus CVK2] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
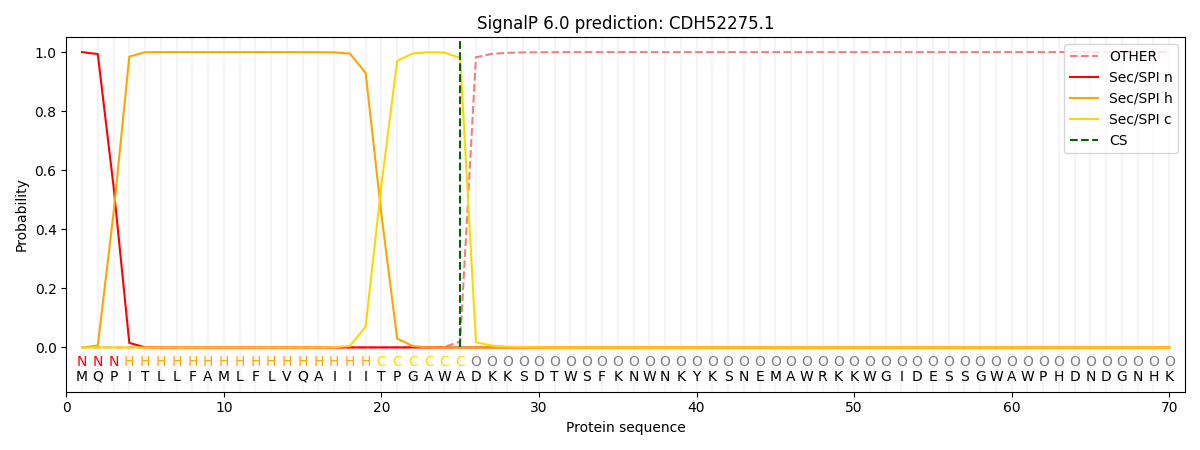
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000214 | 0.999756 | CS pos: 25-26. Pr: 0.9802 |
