You are browsing environment: FUNGIDB
CAZyme Information: CDH50937.1
You are here: Home > Sequence: CDH50937.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
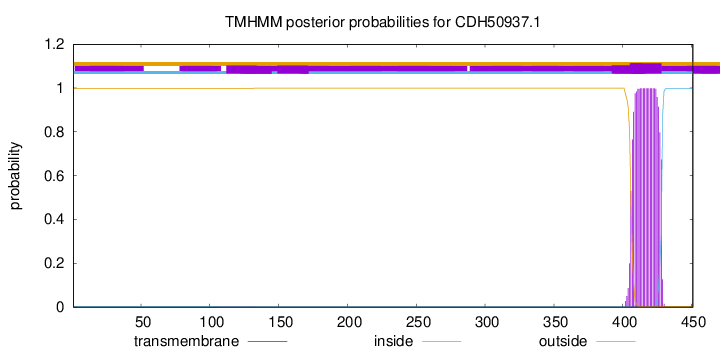
TMHMM annotations
Basic Information help
| Species | Lichtheimia corymbifera | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Mucoromycota; Mucoromycetes; ; Lichtheimiaceae; Lichtheimia; Lichtheimia corymbifera | |||||||||||
| CAZyme ID | CDH50937.1 | |||||||||||
| CAZy Family | GH15 | |||||||||||
| CAZyme Description | alpha-( )-fucosyltransferase 11 | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 2.4.1.152:1 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT10 | 8 | 320 | 2.3e-67 | 0.8443804034582133 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 395683 | Glyco_transf_10 | 5.60e-45 | 148 | 319 | 2 | 157 | Glycosyltransferase family 10 (fucosyltransferase) C-term. This is the C-terminal domain of a family of fucosyltransferases. This enzyme transfers fucose from GDP-Fucose to GlcNAc in an alpha1,3 linkage. This family is known as glycosyltransferase family 10. The C-terminal domain is the likely binding-region for ADP (manuscript in publication). |
| 407214 | Glyco_tran_10_N | 5.37e-04 | 7 | 93 | 2 | 79 | Fucosyltransferase, N-terminal. This is the N-terminal domain of a family of fucosyltransferases. This enzyme transfers fucose from GDP-Fucose to GlcNAc in an alpha1,3 linkage. This family is known as glycosyltransferase family 10. The N-terminal domain is the likely binding-region for the fucose-like substrate (manuscript in publication). |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.11e-155 | 1 | 216 | 1 | 216 | |
| 1.38e-118 | 89 | 435 | 2 | 341 | |
| 8.20e-35 | 79 | 282 | 6 | 225 | |
| 2.01e-34 | 149 | 335 | 10 | 210 | |
| 2.93e-34 | 64 | 335 | 120 | 409 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.07e-06 | 193 | 279 | 223 | 308 | Crystal Structure of alpha1,3-Fucosyltransferase [Helicobacter pylori],2NZW_B Crystal Structure of alpha1,3-Fucosyltransferase [Helicobacter pylori],2NZW_C Crystal Structure of alpha1,3-Fucosyltransferase [Helicobacter pylori],2NZX_A Crystal Structure of alpha1,3-Fucosyltransferase with GDP [Helicobacter pylori],2NZX_B Crystal Structure of alpha1,3-Fucosyltransferase with GDP [Helicobacter pylori],2NZX_C Crystal Structure of alpha1,3-Fucosyltransferase with GDP [Helicobacter pylori],2NZY_A Crystal Structure of alpha1,3-Fucosyltransferase with GDP-fucose [Helicobacter pylori],2NZY_B Crystal Structure of alpha1,3-Fucosyltransferase with GDP-fucose [Helicobacter pylori],2NZY_C Crystal Structure of alpha1,3-Fucosyltransferase with GDP-fucose [Helicobacter pylori] |
|
| 1.20e-06 | 193 | 279 | 223 | 308 | Crystal Structure of alpha1,3-Fucosyltransferase [Helicobacter pylori],5ZOI_B Crystal Structure of alpha1,3-Fucosyltransferase [Helicobacter pylori] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6.45e-35 | 64 | 335 | 120 | 409 | Alpha-(1,3)-fucosyltransferase 11 OS=Homo sapiens OX=9606 GN=FUT11 PE=1 SV=1 |
|
| 1.61e-34 | 64 | 335 | 116 | 409 | Alpha-(1,3)-fucosyltransferase 11 OS=Mus musculus OX=10090 GN=Fut11 PE=1 SV=1 |
|
| 1.96e-34 | 17 | 318 | 92 | 404 | Alpha-(1,3)-fucosyltransferase 11 OS=Gallus gallus OX=9031 GN=FUT11 PE=2 SV=1 |
|
| 8.32e-34 | 64 | 335 | 121 | 410 | Alpha-(1,3)-fucosyltransferase 11 OS=Rattus norvegicus OX=10116 GN=Fut11 PE=2 SV=1 |
|
| 1.70e-33 | 62 | 283 | 125 | 362 | Alpha-(1,3)-fucosyltransferase 11 OS=Takifugu rubripes OX=31033 GN=fut11 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
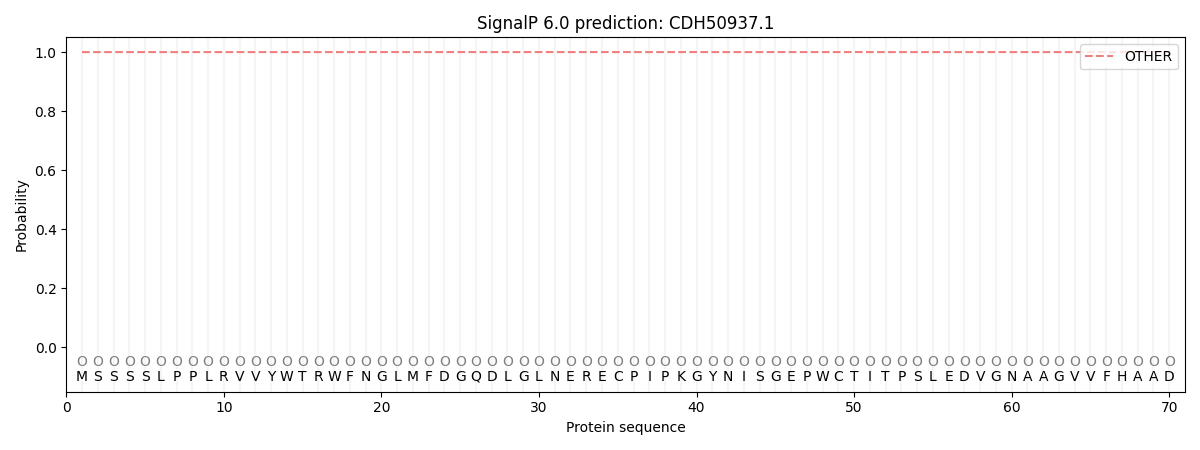
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000036 | 0.000008 |