You are browsing environment: FUNGIDB
CAZyme Information: CDH50470.1
Basic Information
help
| Species |
Lichtheimia corymbifera
|
| Lineage |
Mucoromycota; Mucoromycetes; ; Lichtheimiaceae; Lichtheimia; Lichtheimia corymbifera
|
| CAZyme ID |
CDH50470.1
|
| CAZy Family |
CE8 |
| CAZyme Description |
hypothetical protein RO3G_10121
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 363 |
CBTN010000006|CGC3 |
41755.52 |
6.0062 |
|
| Genome Property |
| Genome Version/Assembly ID |
Genes |
Strain NCBI Taxon ID |
Non Protein Coding Genes |
Protein Coding Genes |
| FungiDB-61_LcorymbiferaJMRCFSU9682 |
12379 |
1263082 |
97 |
12282
|
|
| Gene Location |
No EC number prediction in CDH50470.1.
| Family |
Start |
End |
Evalue |
family coverage |
| GT77 |
149 |
352 |
3.8e-21 |
0.9629629629629629 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| 367480
|
Nucleotid_trans |
7.78e-11 |
149 |
353 |
5 |
205 |
Nucleotide-diphospho-sugar transferase. Proteins in this family have been been predicted to be nucleotide-diphospho-sugar transferases. |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
| 3.34e-225 |
1 |
363 |
1 |
356 |
| 8.72e-144 |
76 |
363 |
81 |
368 |
| 1.99e-74 |
94 |
363 |
67 |
333 |
| 2.19e-71 |
94 |
363 |
105 |
370 |
| 3.11e-70 |
8 |
363 |
10 |
353 |
CDH50470.1 has no PDB hit.
CDH50470.1 has no Swissprot hit.
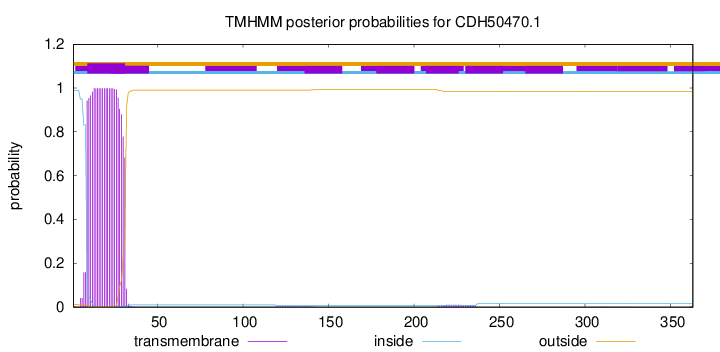
This protein is predicted as OTHER

| Other |
SP_Sec_SPI |
CS Position |
| 0.999539 |
0.000492 |
|