You are browsing environment: FUNGIDB
CDH49253.1
Basic Information
help
Species
Lichtheimia corymbifera
Lineage
Mucoromycota; Mucoromycetes; ; Lichtheimiaceae; Lichtheimia; Lichtheimia corymbifera
CAZyme ID
CDH49253.1
CAZy Family
CE16
CAZyme Description
polysaccharide lyase family 14 protein
CAZyme Property
Protein Length
CGC
Molecular Weight
Isoelectric Point
285
CBTN010000003|CGC3 30138.59
6.3236
Genome Property
Genome Version/Assembly ID
Genes
Strain NCBI Taxon ID
Non Protein Coding Genes
Protein Coding Genes
FungiDB-61_LcorymbiferaJMRCFSU9682
12379
1263082
97
12282
Gene Location
No EC number prediction in CDH49253.1.
Family
Start
End
Evalue
family coverage
PL14
72
281
1.4e-60
0.9809523809523809
CDH49253.1 has no CDD domain.
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
4.70e-164
19
285
16
282
2.60e-130
7
284
7
297
4.29e-100
22
285
37
299
4.55e-68
23
285
48
323
8.68e-53
66
285
3
220
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
Description
1.46e-34
48
282
37
270
Crystal structure of the marine PL-14 alginate lyase from Aplysia kurodai [Aplysia kurodai],5GMT_B Crystal structure of the marine PL-14 alginate lyase from Aplysia kurodai [Aplysia kurodai]
more
CDH49253.1 has no Swissprot hit.
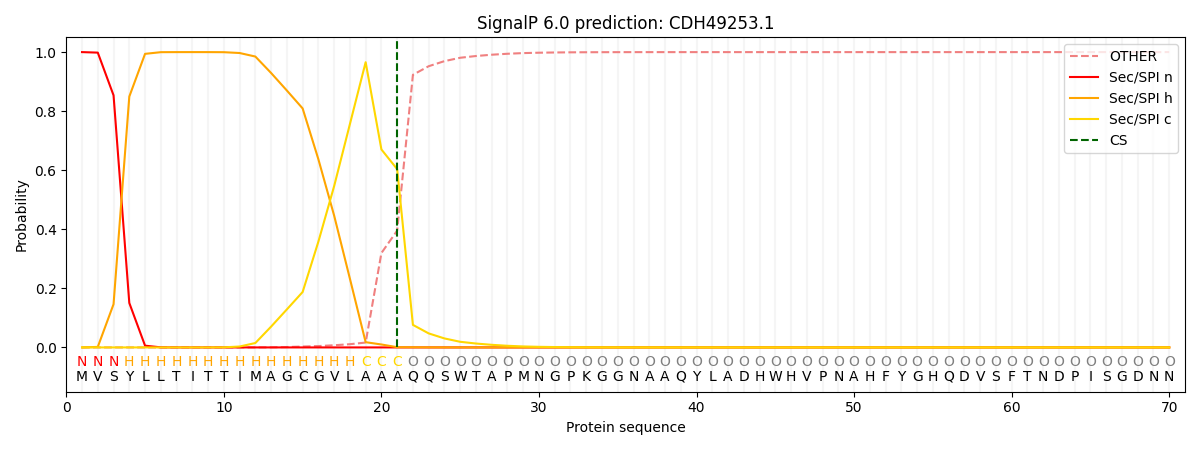
This protein is predicted as SP
Other
SP_Sec_SPI
CS Position
0.000285
0.999680
CS pos: 21-22. Pr: 0.6036