You are browsing environment: FUNGIDB
CAZyme Information: CCG81942.1
You are here: Home > Sequence: CCG81942.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Taphrina deformans | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Taphrinomycetes; ; Taphrinaceae; Taphrina; Taphrina deformans | |||||||||||
| CAZyme ID | CCG81942.1 | |||||||||||
| CAZy Family | GH13|GH13 | |||||||||||
| CAZyme Description | Probable glucan endo-1,3-beta-glucosidase eglC | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH17 | 152 | 404 | 3.5e-19 | 0.9163987138263665 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 227625 | Scw11 | 5.40e-29 | 120 | 394 | 46 | 300 | Exo-beta-1,3-glucanase, GH17 family [Carbohydrate transport and metabolism]. |
| 381626 | GH113_mannanase-like | 0.004 | 295 | 351 | 181 | 240 | Glycoside hydrolase family 113 beta-1,4-mannanase and similar proteins. Mannan endo-1,4-beta mannosidase (E.C 3.2.1.78) randomly cleaves (1->4)-beta-D-mannosidic linkages in mannans, galactomannans and glucomannans and is also called beta-1,4-mannanase, endo-1,4-beta-mannanase, endo-beta-1,4-mannase, beta-mannanase B, beta-1, 4-mannan 4-mannanohydrolase, endo-beta-mannanase, beta-D-mannanase, 1,4-beta-D-mannan mannanohydrolase, and 4-beta-D-mannan mannanohydrolase. (1->4)-beta-linked mannans are polysaccharides with a linear polymer backbone of (1->4)-beta-linked mannose units (in plants and fungi) or alternating mannose and glucose/galactose units (glucomannan in plants and fungi, and galactomannan and galactoglucomannan in plants), such as in the hemicellulose fraction of hard- and softwoods. Complete degradation of mannan requires a series of enzymes, including beta-1,4-mannanase. According to the CAZy database beta-1,4-mannanases are grouped into various glycoside hydrolase (GH) families; GH family 113 beta-1,4-mannanases include mostly bacterial and archaeal sequences. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.56e-115 | 114 | 408 | 12 | 311 | |
| 4.62e-107 | 113 | 407 | 15 | 317 | |
| 5.11e-107 | 112 | 407 | 13 | 314 | |
| 1.59e-106 | 113 | 406 | 14 | 311 | |
| 5.27e-105 | 113 | 407 | 15 | 317 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.49e-14 | 121 | 394 | 39 | 281 | Crystal structure of glycoside hydrolase family 17 beta-1,3-glucanosyltransferase from Rhizomucor miehei [Rhizomucor miehei CAU432] |
|
| 9.00e-14 | 121 | 394 | 39 | 281 | Active-site mutant of Rhizomucor miehei beta-1,3-glucanosyltransferase in complex with laminaribiose [Rhizomucor miehei CAU432],4WTS_A Active-site mutant of Rhizomucor miehei beta-1,3-glucanosyltransferase in complex with laminaritriose [Rhizomucor miehei CAU432] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.87e-102 | 113 | 409 | 14 | 313 | Probable glucan endo-1,3-beta-glucosidase eglC OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=eglC PE=3 SV=1 |
|
| 1.56e-101 | 113 | 406 | 14 | 310 | Probable glucan endo-1,3-beta-glucosidase eglC OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=eglC PE=3 SV=1 |
|
| 3.74e-101 | 113 | 409 | 14 | 313 | Probable glucan endo-1,3-beta-glucosidase eglC OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=eglC PE=3 SV=1 |
|
| 4.21e-101 | 113 | 409 | 14 | 313 | Probable glucan endo-1,3-beta-glucosidase eglC OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=eglC PE=3 SV=1 |
|
| 6.61e-98 | 113 | 409 | 15 | 314 | Probable glucan endo-1,3-beta-glucosidase eglC OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=eglC PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
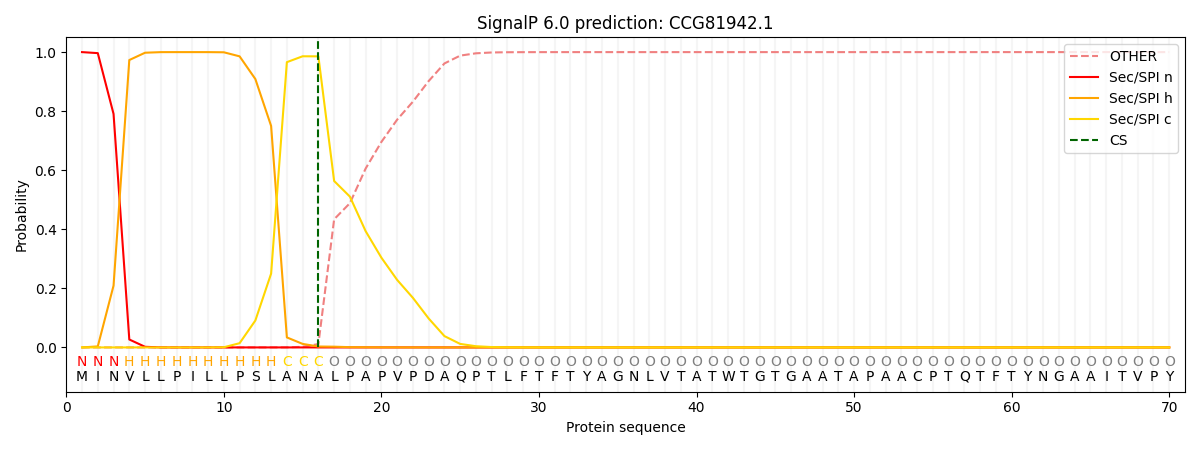
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000220 | 0.999749 | CS pos: 16-17. Pr: 0.9856 |
