You are browsing environment: FUNGIDB
CAZyme Information: CCE32652.1
You are here: Home > Sequence: CCE32652.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Claviceps purpurea | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Clavicipitaceae; Claviceps; Claviceps purpurea | |||||||||||
| CAZyme ID | CCE32652.1 | |||||||||||
| CAZy Family | GH92 | |||||||||||
| CAZyme Description | Chitinase [Source:UniProtKB/TrEMBL;Acc:M1VX99] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.14:1 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH18 | 12 | 270 | 5.2e-25 | 0.7128378378378378 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 212538 | Agmatinase_PAH | 3.09e-138 | 457 | 783 | 1 | 288 | Agmatinase-like family includes proclavaminic acid amidinohydrolase. This agmatinase subfamily contains bacterial and fungal/metazoan enzymes, including proclavaminic acid amidinohydrolase (PAH, EC 3.5.3.22) and Pseudomonas aeruginosa guanidinobutyrase (GbuA) and guanidinopropionase (GpuA). PAH hydrolyzes amidinoproclavaminate to yield proclavaminate and urea in clavulanic acid biosynthesis. Clavulanic acid is an effective inhibitor of beta-lactamases and is used in combination with amoxicillin to prevent the beta-lactam rings of the antibiotic from hydrolysis and, thus keeping the antibiotic biologically active. GbuA hydrolyzes 4-guanidinobutyrate (4-GB) into 4-aminobutyrate and urea while GpuA hydrolyzes 3-guanidinopropionate (3-GP) into beta-alanine and urea. Mutation studies show that significant variations in two active site loops in these two enzymes may be important for substrate specificity. This subfamily belongs to the ureohydrolase superfamily, which includes arginase, agmatinase, proclavaminate amidinohydrolase, and formiminoglutamase. |
| 395395 | Arginase | 8.34e-105 | 478 | 783 | 1 | 270 | Arginase family. |
| 119356 | GH18_hevamine_XipI_class_III | 9.51e-102 | 11 | 308 | 2 | 280 | This conserved domain family includes xylanase inhibitor Xip-I, and the class III plant chitinases such as hevamine, concanavalin B, and PPL2, all of which have a glycosyl hydrolase family 18 (GH18) domain. Hevamine is a class III endochitinase that hydrolyzes the linear polysaccharide chains of chitin and peptidoglycan and is important for defense against pathogenic bacteria and fungi. PPL2 (Parkia platycephala lectin 2) is a class III chitinase from Parkia platycephala seeds that hydrolyzes beta(1-4) glycosidic bonds linking 2-acetoamido-2-deoxy-beta-D-glucopyranose units in chitin. |
| 235011 | PRK02190 | 2.76e-78 | 478 | 783 | 28 | 291 | agmatinase; Provisional |
| 223089 | SpeB | 8.65e-75 | 457 | 790 | 5 | 304 | Arginase family enzyme [Amino acid transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.33e-168 | 1 | 382 | 22 | 379 | |
| 1.42e-162 | 1 | 312 | 21 | 329 | |
| 3.33e-160 | 2 | 312 | 10 | 315 | |
| 1.50e-159 | 1 | 310 | 22 | 328 | |
| 3.43e-156 | 1 | 311 | 21 | 329 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.18e-67 | 12 | 299 | 8 | 272 | ScCTS1_apo crystal structure [Saccharomyces cerevisiae],2UY3_A ScCTS1_8-chlorotheophylline crystal structure [Saccharomyces cerevisiae],2UY4_A ScCTS1_acetazolamide crystal structure [Saccharomyces cerevisiae],2UY5_A ScCTS1_kinetin crystal structure [Saccharomyces cerevisiae],4TXE_A ScCTS1 in complex with compound 5 [Saccharomyces cerevisiae] |
|
| 7.08e-47 | 454 | 784 | 28 | 313 | X-ray crystal structure of a hypothetical Agmatinase from Burkholderia thailandensis [Burkholderia thailandensis E264],4DZ4_B X-ray crystal structure of a hypothetical Agmatinase from Burkholderia thailandensis [Burkholderia thailandensis E264],4DZ4_C X-ray crystal structure of a hypothetical Agmatinase from Burkholderia thailandensis [Burkholderia thailandensis E264],4DZ4_D X-ray crystal structure of a hypothetical Agmatinase from Burkholderia thailandensis [Burkholderia thailandensis E264],4DZ4_E X-ray crystal structure of a hypothetical Agmatinase from Burkholderia thailandensis [Burkholderia thailandensis E264],4DZ4_F X-ray crystal structure of a hypothetical Agmatinase from Burkholderia thailandensis [Burkholderia thailandensis E264] |
|
| 1.68e-45 | 461 | 782 | 12 | 304 | Proclavaminate Amidino Hydrolase From Streptomyces Clavuligerus [Streptomyces clavuligerus],1GQ6_B Proclavaminate Amidino Hydrolase From Streptomyces Clavuligerus [Streptomyces clavuligerus],1GQ6_C Proclavaminate Amidino Hydrolase From Streptomyces Clavuligerus [Streptomyces clavuligerus],1GQ7_A PROCLAVAMINATE AMIDINO HYDROLASE FROM STREPTOMYCES CLAVULIGERUS [Streptomyces clavuligerus],1GQ7_B PROCLAVAMINATE AMIDINO HYDROLASE FROM STREPTOMYCES CLAVULIGERUS [Streptomyces clavuligerus],1GQ7_C PROCLAVAMINATE AMIDINO HYDROLASE FROM STREPTOMYCES CLAVULIGERUS [Streptomyces clavuligerus],1GQ7_D PROCLAVAMINATE AMIDINO HYDROLASE FROM STREPTOMYCES CLAVULIGERUS [Streptomyces clavuligerus],1GQ7_E PROCLAVAMINATE AMIDINO HYDROLASE FROM STREPTOMYCES CLAVULIGERUS [Streptomyces clavuligerus],1GQ7_F PROCLAVAMINATE AMIDINO HYDROLASE FROM STREPTOMYCES CLAVULIGERUS [Streptomyces clavuligerus] |
|
| 2.53e-44 | 12 | 299 | 3 | 292 | A. fumigatus chitinase A1 phenyl-methylguanylurea complex [Aspergillus fumigatus],2XVN_B A. fumigatus chitinase A1 phenyl-methylguanylurea complex [Aspergillus fumigatus],2XVN_C A. fumigatus chitinase A1 phenyl-methylguanylurea complex [Aspergillus fumigatus] |
|
| 2.60e-44 | 12 | 299 | 4 | 293 | AfChiA1 in complex with compound 1 [Aspergillus fumigatus A1163],4TX6_B AfChiA1 in complex with compound 1 [Aspergillus fumigatus A1163] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.10e-122 | 1 | 310 | 18 | 320 | Endochitinase 33 OS=Trichoderma harzianum OX=5544 GN=chit33 PE=1 SV=1 |
|
| 1.52e-121 | 440 | 790 | 24 | 363 | Guanidinobutyrase OS=Kluyveromyces lactis (strain ATCC 8585 / CBS 2359 / DSM 70799 / NBRC 1267 / NRRL Y-1140 / WM37) OX=284590 GN=GBU1 PE=1 SV=1 |
|
| 3.10e-118 | 448 | 792 | 60 | 408 | Putative agmatinase 2 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=SPBC8E4.03 PE=3 SV=1 |
|
| 1.66e-92 | 444 | 792 | 52 | 394 | Putative agmatinase 3 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=SPAPB24D3.03 PE=3 SV=2 |
|
| 9.96e-88 | 455 | 789 | 52 | 379 | Putative agmatinase 1 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=SPAC11D3.09 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
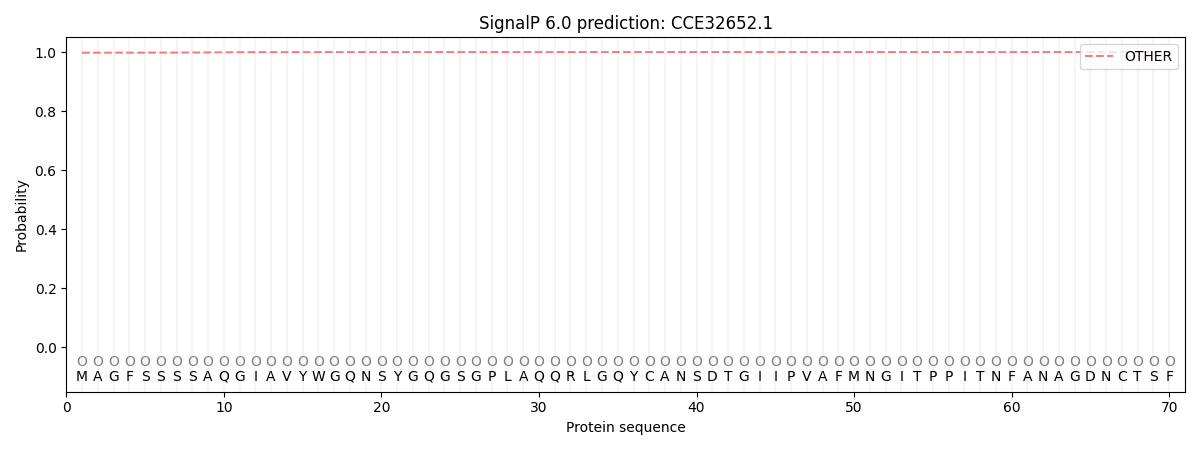
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.998245 | 0.001790 |
