You are browsing environment: FUNGIDB
CAZyme Information: CCE32328.1
You are here: Home > Sequence: CCE32328.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Claviceps purpurea | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Clavicipitaceae; Claviceps; Claviceps purpurea | |||||||||||
| CAZyme ID | CCE32328.1 | |||||||||||
| CAZy Family | GT62 | |||||||||||
| CAZyme Description | Related to beta-glucosidase [Source:UniProtKB/TrEMBL;Acc:M1WDQ5] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 227625 | Scw11 | 3.57e-43 | 252 | 507 | 47 | 299 | Exo-beta-1,3-glucanase, GH17 family [Carbohydrate transport and metabolism]. |
| 163674 | DRE_TIM_metallolyase | 0.006 | 296 | 377 | 117 | 190 | DRE-TIM metallolyase superfamily. The DRE-TIM metallolyase superfamily includes 2-isopropylmalate synthase (IPMS), alpha-isopropylmalate synthase (LeuA), 3-hydroxy-3-methylglutaryl-CoA lyase, homocitrate synthase, citramalate synthase, 4-hydroxy-2-oxovalerate aldolase, re-citrate synthase, transcarboxylase 5S, pyruvate carboxylase, AksA, and FrbC. These members all share a conserved triose-phosphate isomerase (TIM) barrel domain consisting of a core beta(8)-alpha(8) motif with the eight parallel beta strands forming an enclosed barrel surrounded by eight alpha helices. The domain has a catalytic center containing a divalent cation-binding site formed by a cluster of invariant residues that cap the core of the barrel. In addition, the catalytic site includes three invariant residues - an aspartate (D), an arginine (R), and a glutamate (E) - which is the basis for the domain name "DRE-TIM". |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.62e-180 | 1 | 516 | 1 | 572 | |
| 1.78e-148 | 1 | 517 | 1 | 587 | |
| 1.65e-140 | 1 | 514 | 1 | 563 | |
| 3.05e-136 | 1 | 518 | 1 | 569 | |
| 2.50e-134 | 1 | 518 | 1 | 566 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.46e-76 | 245 | 507 | 297 | 553 | Probable beta-glucosidase btgE OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=btgE PE=3 SV=1 |
|
| 1.22e-72 | 252 | 512 | 310 | 564 | Probable beta-glucosidase btgE OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=btgE PE=3 SV=2 |
|
| 1.22e-72 | 252 | 512 | 310 | 564 | Probable beta-glucosidase btgE OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=btgE PE=3 SV=2 |
|
| 9.48e-72 | 252 | 512 | 297 | 551 | Probable beta-glucosidase btgE OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=btgE PE=3 SV=1 |
|
| 1.02e-68 | 1 | 507 | 1 | 559 | Probable beta-glucosidase btgE OS=Aspergillus clavatus (strain ATCC 1007 / CBS 513.65 / DSM 816 / NCTC 3887 / NRRL 1 / QM 1276 / 107) OX=344612 GN=btgE PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
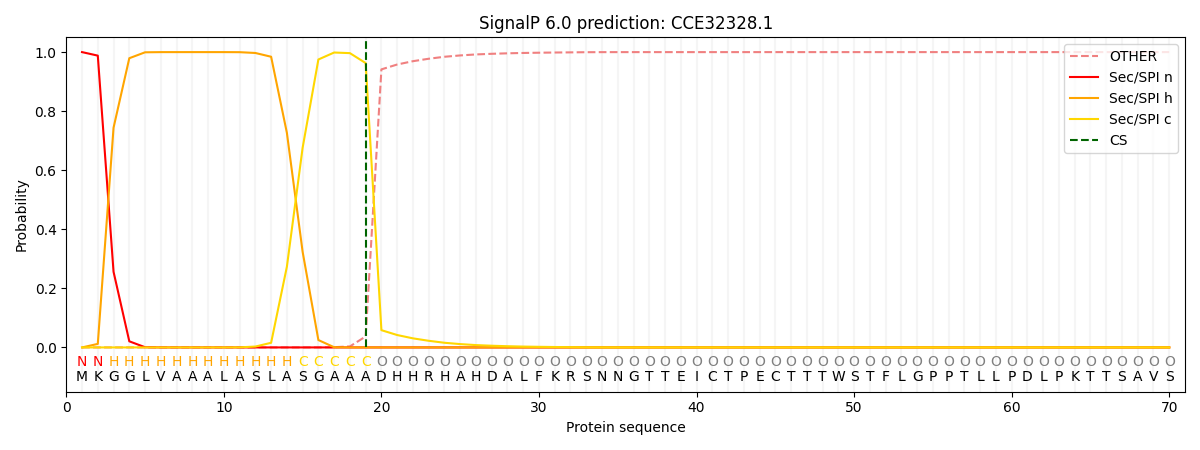
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000324 | 0.999653 | CS pos: 19-20. Pr: 0.9629 |
