You are browsing environment: FUNGIDB
CAZyme Information: CCE31512.1
You are here: Home > Sequence: CCE31512.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Claviceps purpurea | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Clavicipitaceae; Claviceps; Claviceps purpurea | |||||||||||
| CAZyme ID | CCE31512.1 | |||||||||||
| CAZy Family | GT58 | |||||||||||
| CAZyme Description | Related to glucan 1,3-beta-glucosidase [Source:UniProtKB/TrEMBL;Acc:M1VWM1] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 227625 | Scw11 | 1.15e-20 | 11 | 282 | 39 | 280 | Exo-beta-1,3-glucanase, GH17 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.07e-195 | 1 | 329 | 1 | 330 | |
| 3.29e-167 | 5 | 375 | 4 | 367 | |
| 3.56e-165 | 1 | 336 | 1 | 339 | |
| 1.09e-159 | 5 | 312 | 7 | 314 | |
| 2.53e-145 | 3 | 332 | 5 | 330 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.28e-19 | 71 | 306 | 78 | 290 | Crystal structure of glycoside hydrolase family 17 beta-1,3-glucanosyltransferase from Rhizomucor miehei [Rhizomucor miehei CAU432] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.22e-116 | 3 | 312 | 4 | 310 | Probable glucan endo-1,3-beta-glucosidase eglC OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=eglC PE=3 SV=1 |
|
| 1.39e-116 | 3 | 312 | 4 | 310 | Probable glucan endo-1,3-beta-glucosidase eglC OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=eglC PE=3 SV=1 |
|
| 1.18e-115 | 3 | 324 | 4 | 321 | Probable glucan endo-1,3-beta-glucosidase eglC OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=eglC PE=3 SV=1 |
|
| 1.25e-114 | 3 | 326 | 4 | 324 | Probable glucan endo-1,3-beta-glucosidase eglC OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=eglC PE=3 SV=1 |
|
| 7.62e-114 | 3 | 328 | 4 | 327 | Probable glucan endo-1,3-beta-glucosidase eglC OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=eglC PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
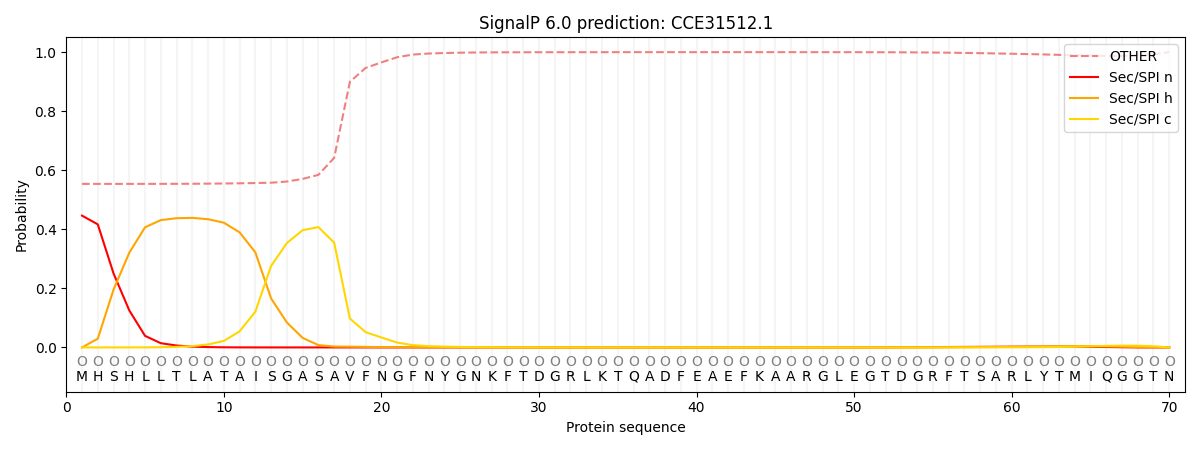
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.574517 | 0.425470 |
