You are browsing environment: FUNGIDB
CAZyme Information: CCE26725.1
You are here: Home > Sequence: CCE26725.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Claviceps purpurea | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Clavicipitaceae; Claviceps; Claviceps purpurea | |||||||||||
| CAZyme ID | CCE26725.1 | |||||||||||
| CAZy Family | AA1 | |||||||||||
| CAZyme Description | Asparagine-linked glycosylation protein 10 [Source:UniProtKB/TrEMBL;Acc:M1W1E5] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT59 | 88 | 366 | 3.6e-75 | 0.6732673267326733 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 398537 | DIE2_ALG10 | 7.26e-119 | 88 | 605 | 7 | 383 | DIE2/ALG10 family. The ALG10 protein from Saccharomyces cerevisiae encodes the alpha-1,2 glucosyltransferase of the endoplasmic reticulum. This protein has been characterized in rat as potassium channel regulator 1. |
| 401668 | QCR10 | 4.33e-12 | 26 | 87 | 1 | 61 | Ubiquinol-cytochrome-c reductase complex subunit (QCR10). The QCR10 family of proteins are a component of the ubiquinol-cytochrome c reductase complex (also known as complex III or cytochrome b-c1 complex). This complex is located on the inner mitochondrial membrane and it couples electron transfer from ubiquinol to cytochrome. This subunit (QCR10) is required for stable association of the iron-sulfur protein with the complex. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 9.55e-255 | 78 | 631 | 56 | 663 | |
| 3.09e-233 | 72 | 626 | 48 | 661 | |
| 3.20e-200 | 76 | 618 | 51 | 644 | |
| 3.20e-200 | 76 | 618 | 51 | 644 | |
| 3.20e-200 | 76 | 618 | 51 | 644 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 8.53e-197 | 76 | 623 | 69 | 658 | Dol-P-Glc:Glc(2)Man(9)GlcNAc(2)-PP-Dol alpha-1,2-glucosyltransferase OS=Gibberella zeae (strain ATCC MYA-4620 / CBS 123657 / FGSC 9075 / NRRL 31084 / PH-1) OX=229533 GN=ALG10 PE=3 SV=1 |
|
| 3.43e-139 | 88 | 612 | 75 | 672 | Dol-P-Glc:Glc(2)Man(9)GlcNAc(2)-PP-Dol alpha-1,2-glucosyltransferase OS=Neurospora crassa (strain ATCC 24698 / 74-OR23-1A / CBS 708.71 / DSM 1257 / FGSC 987) OX=367110 GN=alg-10 PE=3 SV=3 |
|
| 5.39e-121 | 88 | 614 | 74 | 594 | Dol-P-Glc:Glc(2)Man(9)GlcNAc(2)-PP-Dol alpha-1,2-glucosyltransferase OS=Magnaporthe oryzae (strain 70-15 / ATCC MYA-4617 / FGSC 8958) OX=242507 GN=ALG10 PE=3 SV=1 |
|
| 7.06e-67 | 88 | 613 | 41 | 549 | Dol-P-Glc:Glc(2)Man(9)GlcNAc(2)-PP-Dol alpha-1,2-glucosyltransferase OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=alg10 PE=3 SV=1 |
|
| 2.42e-62 | 88 | 613 | 42 | 552 | Dol-P-Glc:Glc(2)Man(9)GlcNAc(2)-PP-Dol alpha-1,2-glucosyltransferase OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=alg10 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
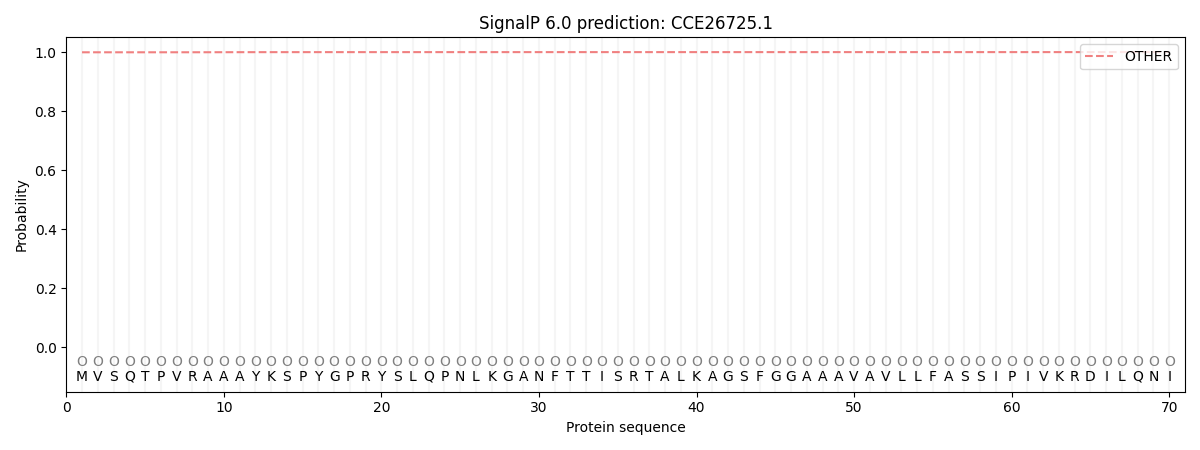
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.999559 | 0.000434 |
TMHMM Annotations download full data without filtering help
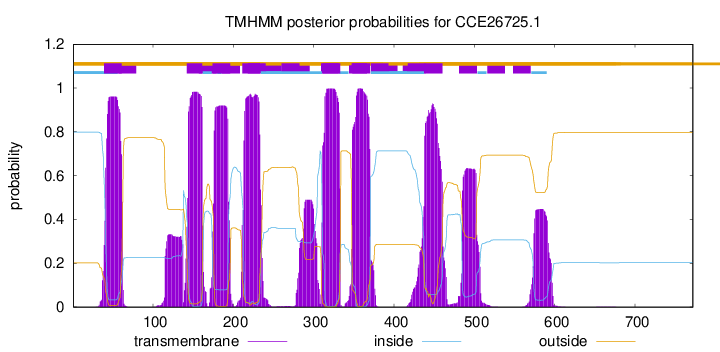
| Start | End |
|---|---|
| 39 | 61 |
| 142 | 161 |
| 174 | 196 |
| 211 | 233 |
| 311 | 333 |
| 348 | 370 |
| 438 | 460 |
