You are browsing environment: FUNGIDB
CAZyme Information: CC1G_15414-t26_1-p1
You are here: Home > Sequence: CC1G_15414-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Coprinopsis cinerea | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Basidiomycota; Agaricomycetes; ; Psathyrellaceae; Coprinopsis; Coprinopsis cinerea | |||||||||||
| CAZyme ID | CC1G_15414-t26_1-p1 | |||||||||||
| CAZy Family | PL35 | |||||||||||
| CAZyme Description | glucose dehydrogenase short protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA3 | 33 | 597 | 1.4e-158 | 0.9947183098591549 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 225186 | BetA | 3.23e-95 | 30 | 596 | 4 | 534 | Choline dehydrogenase or related flavoprotein [Lipid transport and metabolism, General function prediction only]. |
| 235000 | PRK02106 | 1.05e-91 | 33 | 596 | 5 | 532 | choline dehydrogenase; Validated |
| 366272 | GMC_oxred_N | 6.91e-32 | 108 | 355 | 15 | 218 | GMC oxidoreductase. This family of proteins bind FAD as a cofactor. |
| 398739 | GMC_oxred_C | 3.03e-30 | 456 | 591 | 2 | 143 | GMC oxidoreductase. This domain found associated with pfam00732. |
| 215420 | PLN02785 | 1.27e-24 | 34 | 584 | 56 | 565 | Protein HOTHEAD |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 3.51e-143 | 19 | 600 | 19 | 602 | |
| 7.29e-140 | 10 | 596 | 11 | 590 | |
| 6.50e-138 | 14 | 596 | 15 | 590 | |
| 6.50e-138 | 14 | 596 | 15 | 590 | |
| 1.30e-137 | 10 | 596 | 11 | 590 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.19e-140 | 33 | 596 | 1 | 562 | Crystal structure of aryl-alcohol oxidase from Pleurotus eryngii in complex with p-anisic acid [Pleurotus eryngii] |
|
| 1.94e-139 | 33 | 596 | 2 | 563 | Crystal structure of aryl-alcohol-oxidase from Pleurotus eryingii [Pleurotus eryngii] |
|
| 4.38e-126 | 33 | 600 | 40 | 602 | Crystal structure of pyranose dehydrogenase from Agaricus meleagris, wildtype [Leucoagaricus meleagris] |
|
| 1.96e-61 | 32 | 597 | 4 | 584 | Chain A, FAD-dependent oxidoreductase [Thermochaetoides thermophila DSM 1495],6ZE2_B Chain B, FAD-dependent oxidoreductase [Thermochaetoides thermophila DSM 1495],6ZE3_A Chain A, FAD-dependent oxidoreductase [Thermochaetoides thermophila DSM 1495],6ZE4_A Chain A, FAD-dependent oxidoreductase [Thermochaetoides thermophila DSM 1495],6ZE4_B Chain B, FAD-dependent oxidoreductase [Thermochaetoides thermophila DSM 1495],6ZE5_A Chain A, FAD-dependent oxidoreductase [Thermochaetoides thermophila DSM 1495],6ZE5_B Chain B, FAD-dependent oxidoreductase [Thermochaetoides thermophila DSM 1495],6ZE6_A Chain A, FAD-dependent oxidoreductase [Thermochaetoides thermophila DSM 1495],6ZE6_B Chain B, FAD-dependent oxidoreductase [Thermochaetoides thermophila DSM 1495],6ZE7_A Chain A, FAD-dependent oxidoreductase [Thermochaetoides thermophila DSM 1495],6ZE7_B Chain B, FAD-dependent oxidoreductase [Thermochaetoides thermophila DSM 1495],7AA2_A Chain A, FAD-dependent oxidoreductase [Thermochaetoides thermophila DSM 1495],7AA2_B Chain B, FAD-dependent oxidoreductase [Thermochaetoides thermophila DSM 1495] |
|
| 3.22e-59 | 32 | 597 | 4 | 566 | Crystal structure of Aspergillus flavus FAD glucose dehydrogenase [Aspergillus flavus NRRL3357],4YNU_A Crystal structure of Aspergillus flavus FADGDH in complex with D-glucono-1,5-lactone [Aspergillus flavus NRRL3357] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.78e-129 | 9 | 596 | 9 | 596 | Dehydrogenase eriK OS=Hericium erinaceus OX=91752 GN=eriK PE=3 SV=1 |
|
| 9.60e-127 | 4 | 600 | 11 | 600 | Pyranose dehydrogenase 3 OS=Leucoagaricus meleagris OX=201219 GN=pdh3 PE=2 SV=1 |
|
| 2.25e-125 | 33 | 600 | 40 | 602 | Pyranose dehydrogenase 1 OS=Leucoagaricus meleagris OX=201219 GN=pdh1 PE=1 SV=1 |
|
| 1.73e-122 | 11 | 600 | 17 | 594 | Pyranose dehydrogenase OS=Agaricus bisporus OX=5341 GN=pdh1 PE=1 SV=1 |
|
| 1.40e-121 | 33 | 600 | 36 | 595 | Pyranose dehydrogenase OS=Agaricus campestris OX=56157 GN=pdh1 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
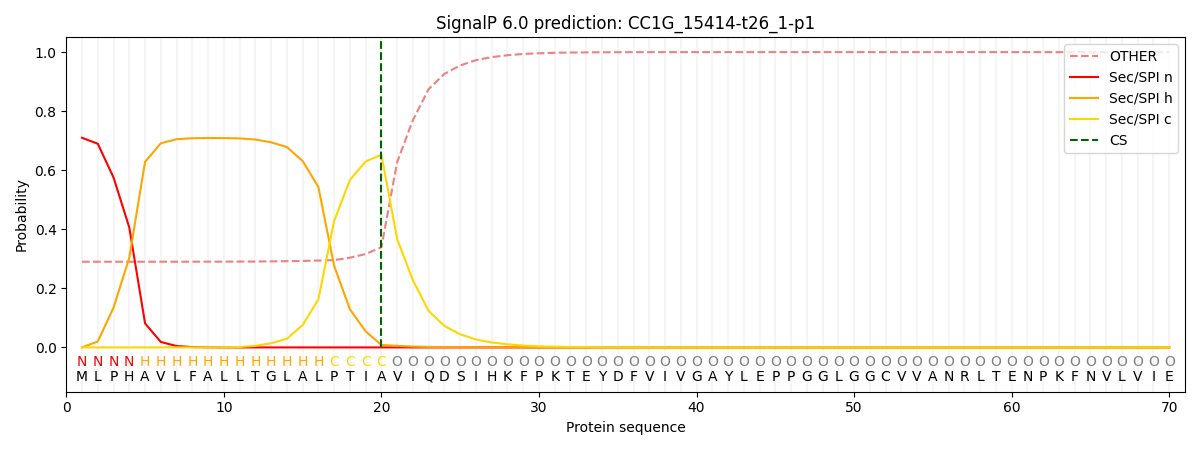
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.298195 | 0.701791 | CS pos: 20-21. Pr: 0.6524 |
