You are browsing environment: FUNGIDB
CAZyme Information: CC1G_11503-t26_1-p1
You are here: Home > Sequence: CC1G_11503-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Coprinopsis cinerea | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Basidiomycota; Agaricomycetes; ; Psathyrellaceae; Coprinopsis; Coprinopsis cinerea | |||||||||||
| CAZyme ID | CC1G_11503-t26_1-p1 | |||||||||||
| CAZy Family | GT2 | |||||||||||
| CAZyme Description | cutinase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE5 | 31 | 198 | 4.7e-43 | 0.9841269841269841 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 395860 | Cutinase | 1.93e-48 | 31 | 199 | 2 | 173 | Cutinase. |
| 238382 | Lipase | 8.79e-05 | 80 | 144 | 1 | 64 | Lipase. Lipases are esterases that can hydrolyze long-chain acyl-triglycerides into di- and monoglycerides, glycerol, and free fatty acids at a water/lipid interface. A typical feature of lipases is "interfacial activation", the process of becoming active at the lipid/water interface, although several examples of lipases have been identified that do not undergo interfacial activation . The active site of a lipase contains a catalytic triad consisting of Ser - His - Asp/Glu, but unlike most serine proteases, the active site is buried inside the structure. A "lid" or "flap" covers the active site, making it inaccessible to solvent and substrates. The lid opens during the process of interfacial activation, allowing the lipid substrate access to the active site. |
| 238287 | Lipase_3 | 0.003 | 96 | 186 | 116 | 201 | Lipase (class 3). Lipases are esterases that can hydrolyze long-chain acyl-triglycerides into di- and monoglycerides, glycerol, and free fatty acids at a water/lipid interface. A typical feature of lipases is "interfacial activation," the process of becoming active at the lipid/water interface, although several examples of lipases have been identified that do not undergo interfacial activation . The active site of a lipase contains a catalytic triad consisting of Ser - His - Asp/Glu, but unlike most serine proteases, the active site is buried inside the structure. A "lid" or "flap" covers the active site, making it inaccessible to solvent and substrates. The lid opens during the process of interfacial activation, allowing the lipid substrate access to the active site. |
| 369775 | PE-PPE | 0.004 | 68 | 148 | 3 | 91 | PE-PPE domain. This domain is found C terminal to the PE (pfam00934) and PPE (pfam00823) domains. The secondary structure of this domain is predicted to be a mixture of alpha helices and beta strands. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 5.35e-103 | 1 | 199 | 1 | 199 | |
| 6.58e-64 | 1 | 199 | 1 | 200 | |
| 2.46e-54 | 14 | 195 | 16 | 196 | |
| 2.46e-54 | 14 | 195 | 16 | 196 | |
| 9.62e-54 | 14 | 195 | 16 | 195 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7.13e-35 | 25 | 199 | 73 | 247 | Structure of cutinase from Trichoderma reesei in its native form. [Trichoderma reesei QM6a],4PSD_A Structure of Trichoderma reesei cutinase native form. [Trichoderma reesei QM6a],4PSE_A Trichoderma reesei cutinase in complex with a C11Y4 phosphonate inhibitor [Trichoderma reesei QM6a],4PSE_B Trichoderma reesei cutinase in complex with a C11Y4 phosphonate inhibitor [Trichoderma reesei QM6a] |
|
| 4.72e-25 | 25 | 199 | 17 | 198 | Chain A, cutinase [Malbranchea cinnamomea] |
|
| 1.01e-23 | 20 | 188 | 6 | 195 | Chain A, CUTINASE [Fusarium vanettenii],1CUW_B Chain B, CUTINASE [Fusarium vanettenii] |
|
| 7.68e-23 | 20 | 188 | 6 | 195 | Chain A, CUTINASE [Fusarium vanettenii] |
|
| 1.51e-22 | 20 | 188 | 6 | 195 | Chain A, CUTINASE [Fusarium vanettenii] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 9.50e-104 | 1 | 199 | 1 | 199 | Cutinase CUT1 OS=Coprinopsis cinerea OX=5346 GN=CUT1 PE=1 SV=1 |
|
| 4.37e-55 | 14 | 195 | 16 | 196 | Cutinase OS=Botryotinia fuckeliana OX=40559 GN=cutA PE=1 SV=1 |
|
| 1.71e-54 | 14 | 195 | 16 | 195 | Cutinase OS=Monilinia fructicola OX=38448 GN=CUT1 PE=2 SV=1 |
|
| 1.57e-39 | 1 | 198 | 1 | 200 | Cutinase pbc1 OS=Pyrenopeziza brassicae OX=76659 GN=pbc1 PE=1 SV=1 |
|
| 2.71e-36 | 31 | 195 | 60 | 217 | Cutinase 4 OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=cut4 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
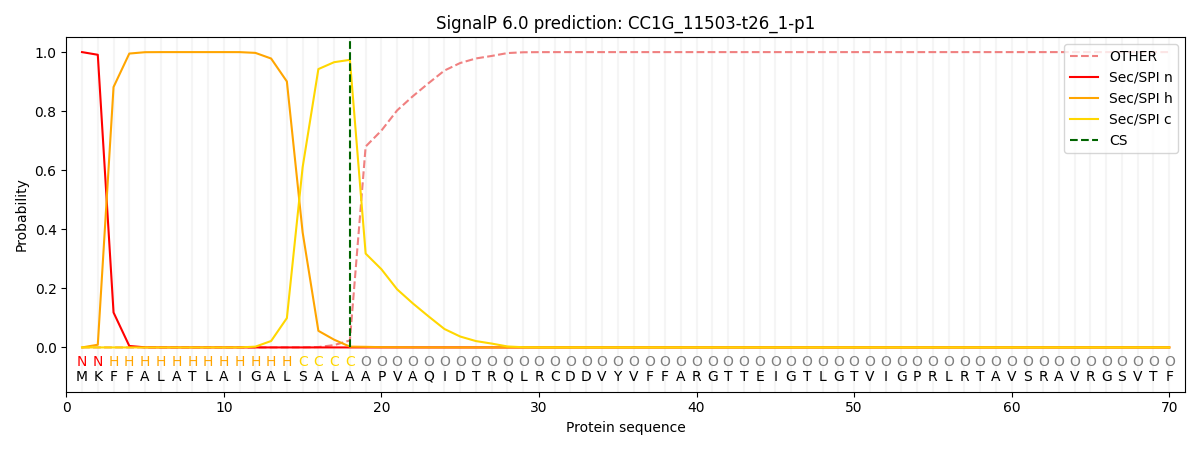
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000236 | 0.999749 | CS pos: 18-19. Pr: 0.9738 |
