You are browsing environment: FUNGIDB
CAZyme Information: CC1G_06647-t26_1-p1
You are here: Home > Sequence: CC1G_06647-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Coprinopsis cinerea | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Basidiomycota; Agaricomycetes; ; Psathyrellaceae; Coprinopsis; Coprinopsis cinerea | |||||||||||
| CAZyme ID | CC1G_06647-t26_1-p1 | |||||||||||
| CAZy Family | GH16 | |||||||||||
| CAZyme Description | mannosyltransferase 1 | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 2.4.1.109:12 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT39 | 83 | 322 | 6.9e-74 | 0.9910313901345291 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 224839 | PMT1 | 0.0 | 79 | 761 | 26 | 698 | Dolichyl-phosphate-mannose--protein O-mannosyl transferase [Posttranslational modification, protein turnover, chaperones]. |
| 406576 | PMT_4TMC | 1.23e-87 | 568 | 754 | 2 | 198 | C-terminal four TMM region of protein-O-mannosyltransferase. PMT_4TMC is the C-terminal four membrane-pass region of protein-O-mannosyltransferases and similar enzymes. |
| 396786 | PMT | 1.21e-82 | 81 | 325 | 1 | 244 | Dolichyl-phosphate-mannose-protein mannosyltransferase. This is a family of Dolichyl-phosphate-mannose-protein mannosyltransferase proteins EC:2.4.1.109. These proteins are responsible for O-linked glycosylation of proteins, they catalyze the reaction:- Dolichyl phosphate D-mannose + protein <=> dolichyl phosphate + O-D-mannosyl-protein. Also in this family is Drosophila rotated abdomen protein which is a putative mannosyltransferase. This family appears to be distantly related to pfam02516 (A Bateman pers. obs.). This family also contains sequences from ArnTs (4-amino-4-deoxy-L-arabinose lipid A transferase). They catalyze the addition of 4-amino-4-deoxy-l-arabinose (l-Ara4N) to the lipid A moiety of the lipopolysaccharide. This is a critical modification enabling bacteria (e.g. Escherichia coli and Salmonella typhimurium) to resist killing by antimicrobial peptides such as polymyxins. Members such as undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase are predicted to have 12 trans-membrane regions. The N-terminal portion of these proteins is hypothesized to have a conserved glycosylation activity which is shared between distantly related oligosaccharyltransferases ArnT and PglB families. |
| 397103 | MIR | 7.81e-39 | 374 | 541 | 13 | 183 | MIR domain. The MIR (protein mannosyltransferase, IP3R and RyR) domain is a domain that may have a ligand transferase function. |
| 197746 | MIR | 5.20e-14 | 498 | 546 | 8 | 57 | Domain in ryanodine and inositol trisphosphate receptors and protein O-mannosyltransferases. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 10 | 828 | 5 | 820 | |
| 0.0 | 10 | 828 | 5 | 820 | |
| 0.0 | 10 | 828 | 5 | 820 | |
| 0.0 | 10 | 828 | 5 | 798 | |
| 0.0 | 1 | 833 | 1 | 882 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6.06e-199 | 72 | 828 | 45 | 808 | Structure of S. cerevisiae protein O-mannosyltransferase Pmt1-Pmt2 complex bound to the sugar donor and a peptide acceptor [Saccharomyces cerevisiae W303],6P2R_A Structure of S. cerevisiae protein O-mannosyltransferase Pmt1-Pmt2 complex bound to the sugar donor [Saccharomyces cerevisiae W303] |
|
| 2.27e-140 | 39 | 759 | 35 | 754 | Structure of S. cerevisiae protein O-mannosyltransferase Pmt1-Pmt2 complex bound to the sugar donor and a peptide acceptor [Saccharomyces cerevisiae W303],6P2R_B Structure of S. cerevisiae protein O-mannosyltransferase Pmt1-Pmt2 complex bound to the sugar donor [Saccharomyces cerevisiae W303] |
|
| 4.54e-37 | 354 | 545 | 5 | 192 | Crystal structure of the MIR domain (aa 337-532) of the S. cerevisiae mannosyltransferase Pmt2 [Saccharomyces cerevisiae] |
|
| 7.63e-37 | 354 | 545 | 22 | 209 | Structure of the Pmt2-MIR domain with bound ligands [Saccharomyces cerevisiae] |
|
| 1.86e-34 | 354 | 552 | 14 | 209 | Structure of the Pmt3-MIR domain with bound ligands [Saccharomyces cerevisiae],6ZQQ_B Structure of the Pmt3-MIR domain with bound ligands [Saccharomyces cerevisiae],6ZQQ_C Structure of the Pmt3-MIR domain with bound ligands [Saccharomyces cerevisiae],6ZQQ_D Structure of the Pmt3-MIR domain with bound ligands [Saccharomyces cerevisiae] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 8.60e-212 | 71 | 785 | 60 | 774 | Dolichyl-phosphate-mannose--protein mannosyltransferase 1 OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=PMT1 PE=2 SV=1 |
|
| 3.12e-198 | 72 | 828 | 45 | 808 | Dolichyl-phosphate-mannose--protein mannosyltransferase 1 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=PMT1 PE=1 SV=1 |
|
| 2.91e-163 | 72 | 771 | 40 | 738 | Dolichyl-phosphate-mannose--protein mannosyltransferase 5 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=PMT5 PE=1 SV=1 |
|
| 7.21e-143 | 37 | 758 | 16 | 750 | Protein O-mannosyl-transferase 2 OS=Homo sapiens OX=9606 GN=POMT2 PE=1 SV=2 |
|
| 5.47e-141 | 74 | 758 | 125 | 820 | Protein O-mannosyl-transferase 2 OS=Mus musculus OX=10090 GN=Pomt2 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000041 | 0.000026 |
TMHMM Annotations download full data without filtering help
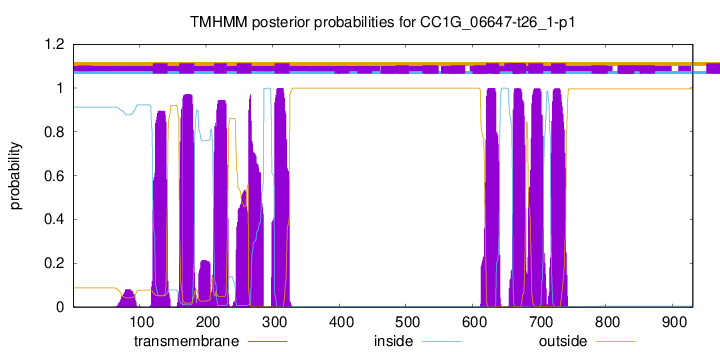
| Start | End |
|---|---|
| 120 | 142 |
| 160 | 182 |
| 212 | 231 |
| 246 | 268 |
| 303 | 325 |
| 621 | 640 |
| 661 | 680 |
| 684 | 706 |
| 718 | 740 |
