You are browsing environment: FUNGIDB
CAZyme Information: CC1G_05972-t26_1-p1
You are here: Home > Sequence: CC1G_05972-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Coprinopsis cinerea | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Basidiomycota; Agaricomycetes; ; Psathyrellaceae; Coprinopsis; Coprinopsis cinerea | |||||||||||
| CAZyme ID | CC1G_05972-t26_1-p1 | |||||||||||
| CAZy Family | GH11 | |||||||||||
| CAZyme Description | laccase 4 | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 1.10.3.2:77 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA1 | 37 | 342 | 1e-147 | 0.9933993399339934 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 259970 | CuRO_3_Tv-LCC_like | 8.13e-80 | 350 | 496 | 1 | 147 | The third cupredoxin domain of the fungal laccases similar to Tv-LCC from Trametes Versicolor. This subfamily of fungal laccases includes Tv-LCC from Trametes versicolor and Rs-LCC2 from plant pathogenic fungus Rhizoctonia solani. Laccase is a blue multi-copper enzyme that catalyzes the oxidation of a variety aromatic - notably phenolic and inorganic substances coupled to the reduction of molecular oxygen to water. It has been implicated in a wide spectrum of biological activities and, in particular, plays a key role in morphogenesis, development and lignin metabolism. Although MCOs have diverse functions, majority of them have three cupredoxin domain repeats that include one mononuclear and one trinuclear copper center. The copper ions are bound in several sites: Type 1, Type 2, and/or Type 3. The ensemble of types 2 and 3 copper is called a trinuclear cluster. MCOs oxidize their substrate by accepting electrons at a mononuclear copper center and transferring them to the active site trinuclear copper center. The cupredoxin domain 3 of 3-domain MCOs contains the Type 1 (T1) copper binding site and part the trinuclear copper binding site, which is located at the interface of domains 1 and 3. |
| 274555 | ascorbase | 1.60e-79 | 32 | 497 | 8 | 526 | L-ascorbate oxidase, plant type. Members of this protein family are the copper-containing enzyme L-ascorbate oxidase (EC 1.10.3.3), also called ascorbase. This family is found in flowering plants, and shows greater sequence similarity to a family of laccases (EC 1.10.3.2) from plants than to other known ascorbate oxidases. |
| 259925 | CuRO_1_Tv-LCC_like | 1.63e-77 | 26 | 150 | 1 | 125 | The first cupredoxin domain of fungal laccases similar to Tv-LCC from Trametes versicolor. This subfamily of fungal laccases includes Tv-LCC from Trametes versicolor and Rs-LCC2 from plant pathogenic fungus Rhizoctonia solani. Laccase is a blue multi-copper enzyme that catalyzes the oxidation of a variety aromatic - notably phenolic and inorganic substances coupled to the reduction of molecular oxygen to water. It has been implicated in a wide spectrum of biological activities and, in particular, plays a key role in morphogenesis, development and lignin metabolism. Although MCOs have diverse functions, majority of them have three cupredoxin domain repeats that include one mononuclear and one trinuclear copper center. The copper ions are bound in several sites: Type 1, Type 2, and/or Type 3. The ensemble of types 2 and 3 copper is called a trinuclear cluster. MCOs oxidize their substrate by accepting electrons at a mononuclear copper center and transferring them to the active site trinuclear copper center. The cupredoxin domain 1 of 3-domain MCOs contains part the trinuclear copper binding site, which is located at the interface of domains 1 and 3. |
| 259949 | CuRO_2_Tv-LCC_like | 1.03e-75 | 166 | 314 | 1 | 151 | The second cupredoxin domain of the fungal laccases similar to Tv-LCC from Trametes versicolor. This subfamily of fungal laccases includes Tv-LCC from Trametes versicolor and Rs-LCC2 from plant pathogenic fungus Rhizoctonia solani. Laccase is a blue multi-copper enzyme that catalyzes the oxidation of a variety aromatic - notably phenolic and inorganic substances coupled to the reduction of molecular oxygen to water. It has been implicated in a wide spectrum of biological activities and, in particular, plays a key role in morphogenesis, development and lignin metabolism. Laccase is a multicopper oxidase (MCO) composed of three cupredoxin domains that include one mononuclear and one trinuclear copper center. The copper ions are bound in several sites: Type 1, Type 2, and/or Type 3. The ensemble of types 2 and 3 copper is called a trinuclear cluster. MCOs oxidize their substrate by accepting electrons at a mononuclear copper center and transferring them to the active site trinuclear copper center. The cupredoxin domain 2 of 3-domain MCOs has lost the ability to bind copper. |
| 177843 | PLN02191 | 5.68e-70 | 28 | 497 | 26 | 549 | L-ascorbate oxidase |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 545 | 1 | 545 | |
| 0.0 | 1 | 545 | 1 | 545 | |
| 2.59e-250 | 3 | 532 | 6 | 537 | |
| 1.05e-249 | 3 | 532 | 6 | 537 | |
| 1.09e-241 | 28 | 528 | 32 | 532 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5.82e-240 | 22 | 526 | 1 | 500 | Chain A, Laccase [Coprinopsis cinerea] |
|
| 1.13e-239 | 23 | 526 | 2 | 500 | Chain A, LACCASE 1 [Coprinopsis cinerea] |
|
| 1.30e-208 | 24 | 521 | 2 | 496 | Chain A, LACCASE 2 [Trametes versicolor] |
|
| 1.35e-207 | 24 | 524 | 2 | 496 | PM1 mutant, 7D5 [Aspergillus oryzae],6H5Y_B PM1 mutant, 7D5 [Aspergillus oryzae] |
|
| 8.31e-207 | 30 | 519 | 8 | 493 | Chain A, Laccase [Lentinus tigrinus],2QT6_B Chain B, Laccase [Lentinus tigrinus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.46e-207 | 24 | 524 | 25 | 527 | Laccase-2 OS=Pleurotus ostreatus OX=5322 GN=POX2 PE=1 SV=1 |
|
| 7.79e-207 | 30 | 524 | 31 | 525 | Laccase-1 OS=Pleurotus ostreatus OX=5322 GN=POX1 PE=2 SV=1 |
|
| 4.46e-206 | 24 | 521 | 22 | 516 | Laccase-2 OS=Trametes villosa OX=47662 GN=LCC2 PE=3 SV=1 |
|
| 1.87e-205 | 24 | 519 | 23 | 515 | Laccase OS=Trametes hirsuta OX=5327 PE=1 SV=1 |
|
| 7.28e-205 | 24 | 521 | 22 | 516 | Laccase-2 OS=Trametes versicolor OX=5325 GN=LCC2 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
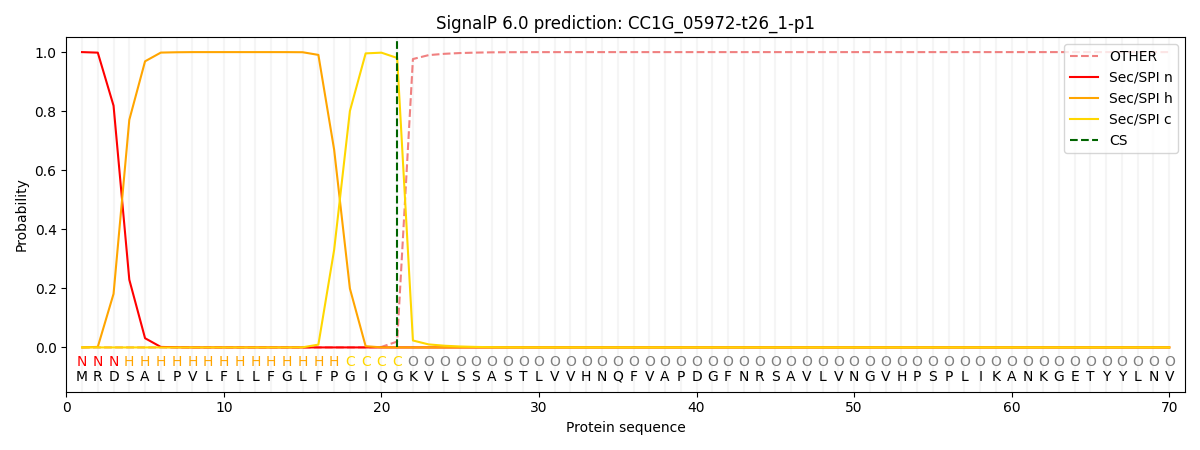
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000232 | 0.999747 | CS pos: 21-22. Pr: 0.9801 |
