You are browsing environment: FUNGIDB
CAZyme Information: CC1G_04177-t26_1-p1
You are here: Home > Sequence: CC1G_04177-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Coprinopsis cinerea | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Basidiomycota; Agaricomycetes; ; Psathyrellaceae; Coprinopsis; Coprinopsis cinerea | |||||||||||
| CAZyme ID | CC1G_04177-t26_1-p1 | |||||||||||
| CAZy Family | CBM21|GH13 | |||||||||||
| CAZyme Description | xylanase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.8:61 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH11 | 50 | 230 | 2.9e-51 | 0.9830508474576272 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 395367 | Glyco_hydro_11 | 1.52e-82 | 50 | 230 | 1 | 175 | Glycosyl hydrolases family 11. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.28e-84 | 1 | 233 | 1 | 229 | |
| 6.65e-74 | 1 | 233 | 1 | 225 | |
| 2.23e-72 | 32 | 233 | 13 | 215 | |
| 4.45e-71 | 40 | 233 | 37 | 232 | |
| 1.81e-69 | 38 | 233 | 31 | 228 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.68e-64 | 42 | 233 | 3 | 190 | Chain A, Protein (endo-1,4-beta-xylanase) [Paecilomyces variotii] |
|
| 3.54e-62 | 42 | 233 | 3 | 190 | ENDO-1,4-BETA-XYLANASE, ROOM TEMPERATURE, PH 4.0 [Thermomyces lanuginosus] |
|
| 2.40e-56 | 47 | 233 | 7 | 188 | Chain A, Endo-1,4-beta-xylanase 2 [Trichoderma reesei] |
|
| 3.29e-56 | 47 | 233 | 6 | 187 | Chain A, Endo-1,4-beta-xylanase 2 [Trichoderma reesei] |
|
| 3.39e-56 | 47 | 233 | 7 | 188 | Crystal Structures of Mutant Endo-beta-1,4-xylanase(Y77F) [Trichoderma reesei RUT C-30],5ZKZ_A Crystal Structures of Mutant Endo-beta-1,4-xylanase II(Y77F) Complexed with Xylotriose [Trichoderma reesei RUT C-30] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.75e-67 | 41 | 233 | 2 | 196 | Endo-1,4-beta-xylanase A OS=Schizophyllum commune OX=5334 GN=XYNA PE=1 SV=1 |
|
| 6.04e-64 | 41 | 233 | 39 | 233 | Endo-1,4-beta-xylanase 1 OS=Leucoagaricus gongylophorus OX=79220 GN=Xyn1 PE=1 SV=1 |
|
| 1.38e-63 | 42 | 233 | 3 | 190 | Endo-1,4-beta-xylanase OS=Byssochlamys spectabilis OX=264951 PE=1 SV=1 |
|
| 2.22e-63 | 38 | 233 | 40 | 229 | Endo-1,4-beta-xylanase 4 OS=Magnaporthe grisea OX=148305 GN=XYL4 PE=3 SV=2 |
|
| 2.22e-63 | 38 | 233 | 40 | 229 | Endo-1,4-beta-xylanase 4 OS=Magnaporthe oryzae (strain 70-15 / ATCC MYA-4617 / FGSC 8958) OX=242507 GN=XYL4 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
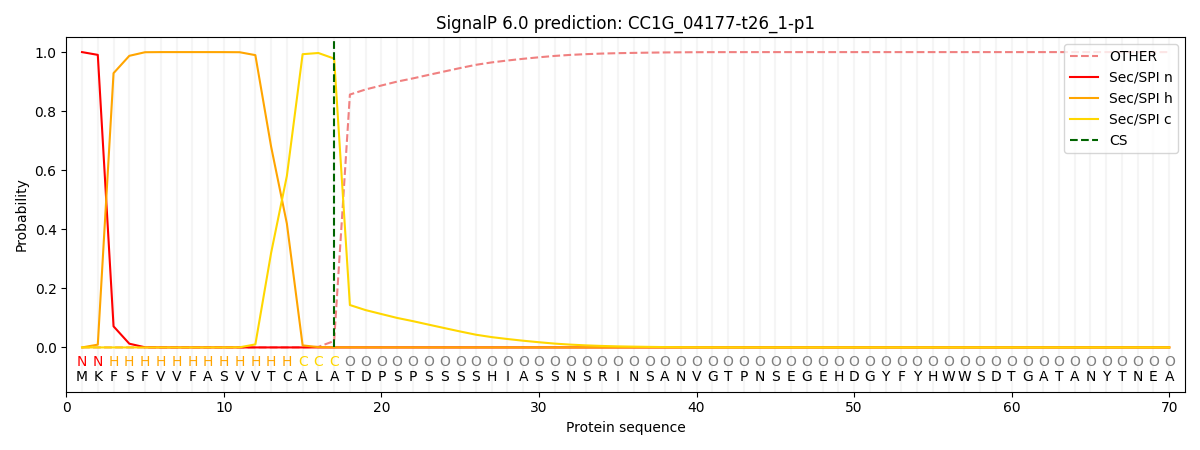
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000258 | 0.999736 | CS pos: 17-18. Pr: 0.9780 |
