You are browsing environment: FUNGIDB
CAZyme Information: CC1G_02937-t26_1-p1
You are here: Home > Sequence: CC1G_02937-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Coprinopsis cinerea | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Basidiomycota; Agaricomycetes; ; Psathyrellaceae; Coprinopsis; Coprinopsis cinerea | |||||||||||
| CAZyme ID | CC1G_02937-t26_1-p1 | |||||||||||
| CAZy Family | AA8|AA3 | |||||||||||
| CAZyme Description | rhamnogalacturonase B | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 4.2.2.23:4 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL4 | 99 | 361 | 6.5e-84 | 0.56312625250501 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 401282 | RhgB_N | 5.64e-46 | 18 | 160 | 1 | 251 | Rhamnogalacturonan lyase B, N-terminal. Members of this family are found in both fungi, bacteria and wood-eating arthropods. The domain is found at the N-terminus of rhamnogalacturonase B, a member of the polysaccharide lyase family 4. The domain adopts a structure consisting of a beta super-sandwich, with eighteen strands in two beta-sheets. The three domains of the whole protein rhamnogalacturonan lyase (RGL4), are involved in the degradation of rhamnogalacturonan-I, RG-I, an important pectic plant cell-wall polysaccharide. The active-site residues are a lysine at position 169 in UniProtKB:Q00019 and a histidine at 229, Lys169 is likely to be a proton abstractor, His229 a proton donor in the mechanism. The substrate is a disaccharide, and RGL4, in contrast to other rhamnogalacturonan hydrolases, cleaves the alpha-1,4 linkages of RG-I between Rha and GalUA through a beta-elimination resulting in a double bond in the nonreducing GalUA residue, and is thus classified as a polysaccharide lyase (PL). |
| 405384 | CBM-like | 1.57e-23 | 253 | 353 | 1 | 114 | Polysaccharide lyase family 4, domain III. CBM-like is domain III of rhamnogalacturonan lyase (RG-lyase). The full-length protein specifically recognizes and cleaves alpha-1,4 glycosidic bonds between l-rhamnose and d-galacturonic acids in the backbone of rhamnogalacturonan-I, a major component of the plant cell wall polysaccharide, pectin. This domain possesses a jelly roll beta-sandwich fold structurally homologous to carbohydrate binding modules (CBMs), and it carries two sulfate ions and a hexa-coordinated calcium ion. |
| 199905 | RGL4_C | 1.13e-17 | 255 | 358 | 1 | 123 | C-terminal domain of rhamnogalacturonan lyase, a family 4 polysaccharide lyase. The rhamnogalacturonan lyase of the polysaccharide lyase family 4 (RGL4) is involved in the degradation of RG (rhamnogalacturonan) type-I, an important pectic plant cell wall polysaccharide, by cleaving the alpha-1,4 glycoside bond between L-rhamnose and D-galacturonic acids in the backbone of RG type-I through a beta-elimination reaction. RGL4 consists of three domains, an N-terminal catalytic domain, a middle domain with a FNIII type fold and a C-terminal domain with a jelly roll fold. Both the middle and the C-terminal domain are putative carbohydrate binding modules. There are two types of RG lyases, which both cleave the alpha-1,4 bonds of the RG-I main chain (RG chain) through the beta-elimination reaction, but belong to two structurally unrelated polysaccharide lyase (PL) families, 4 and 11. |
| 405387 | fn3_3 | 1.72e-12 | 172 | 240 | 3 | 73 | Polysaccharide lyase family 4, domain II. FnIII-like is domain II of rhamnogalacturonan lyase (RG-lyase). The full-length protein specifically recognizes and cleaves alpha-1,4 glycosidic bonds between l-rhamnose and d-galacturonic acids in the backbone of rhamnogalacturonan-I, a major component of the plant cell wall polysaccharide, pectin. This domain displays an immunoglobulin-like or more specifically Fibronectin-III type fold and shows highest structural similarity to the C-terminal beta-sandwich subdomain of the pro-hormone/propeptide processing enzyme carboxypeptidase gp180 from duck. It serves to assist in producing the deep pocket, with domain III, into which the substrate fits. |
| 199904 | RGL4_M | 4.81e-12 | 166 | 243 | 1 | 90 | Middle domain of rhamnogalacturonan lyase, a family 4 polysaccharide lyase. The rhamnogalacturonan lyase of the polysaccharide lyase family 4 (RGL4) is involved in the degradation of RG (rhamnogalacturonan) type-I, an important pectic plant cell wall polysaccharide, by cleaving the alpha-1,4 glycoside bond between L-rhamnose and D-galacturonic acids in the backbone of RG type-I through a beta-elimination reaction. RGL4 consists of three domains, an N-terminal catalytic domain, a middle domain with a FNIII type fold and a C-terminal domain with a jelly roll fold. Both the middle domain represented by this model and the C-terminal domain are putative carbohydrate binding modules. There are two types of RG lyases, which both cleave the alpha-1,4 bonds of the RG-I main chain (RG chain) through the beta-elimination reaction, but belong to two structurally unrelated polysaccharide lyase (PL) families, 4 and 11. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 8.76e-111 | 2 | 358 | 7 | 481 | |
| 1.73e-96 | 12 | 358 | 16 | 472 | |
| 1.68e-94 | 12 | 358 | 16 | 472 | |
| 2.27e-91 | 98 | 352 | 184 | 454 | |
| 3.34e-83 | 11 | 358 | 12 | 480 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5.26e-59 | 104 | 360 | 194 | 467 | Rhamnogalacturonan lyase from Aspergillus aculeatus [Aspergillus aculeatus] |
|
| 5.26e-59 | 104 | 360 | 194 | 467 | Rhamnogalacturonan lyase from Aspergillus aculeatus K150A active site mutant [Aspergillus aculeatus],2XHN_B Rhamnogalacturonan lyase from Aspergillus aculeatus K150A active site mutant [Aspergillus aculeatus],3NJV_A Rhamnogalacturonan lyase from Aspergillus aculeatus K150A substrate complex [Aspergillus aculeatus] |
|
| 1.52e-57 | 104 | 360 | 194 | 467 | Rhamnogalacturonan Lyase from Aspergillus aculeatus mutant H210A [Aspergillus aculeatus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.02e-60 | 14 | 360 | 18 | 487 | Probable rhamnogalacturonate lyase A OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=rglA PE=3 SV=2 |
|
| 3.61e-60 | 98 | 360 | 207 | 486 | Rhamnogalacturonate lyase A OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=rglA PE=2 SV=1 |
|
| 1.42e-59 | 3 | 360 | 7 | 487 | Probable rhamnogalacturonate lyase A OS=Aspergillus clavatus (strain ATCC 1007 / CBS 513.65 / DSM 816 / NCTC 3887 / NRRL 1 / QM 1276 / 107) OX=344612 GN=rglA PE=3 SV=1 |
|
| 3.99e-58 | 104 | 360 | 213 | 486 | Rhamnogalacturonate lyase A OS=Aspergillus aculeatus OX=5053 GN=rglA PE=1 SV=1 |
|
| 4.63e-57 | 104 | 360 | 213 | 486 | Rhamnogalacturonate lyase A (Fragment) OS=Aspergillus niger OX=5061 GN=rglA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
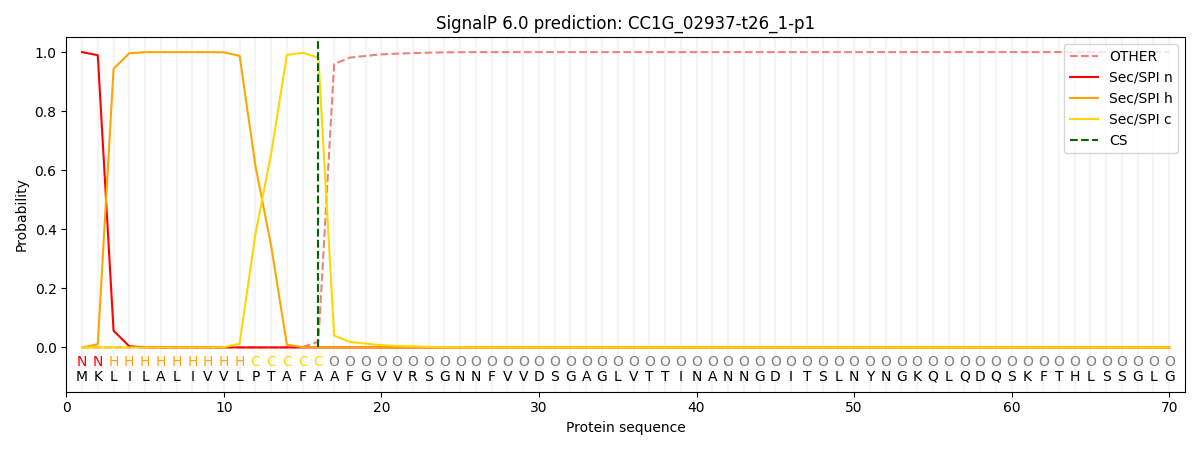
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000213 | 0.999751 | CS pos: 16-17. Pr: 0.9808 |
