You are browsing environment: FUNGIDB
CAZyme Information: CAX43285.1
You are here: Home > Sequence: CAX43285.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Candida dubliniensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Saccharomycetes; ; Debaryomycetaceae; Candida; Candida dubliniensis | |||||||||||
| CAZyme ID | CAX43285.1 | |||||||||||
| CAZy Family | GT2|GT2 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH76 | 76 | 371 | 5.3e-40 | 0.8463687150837989 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 397638 | Glyco_hydro_76 | 1.58e-25 | 115 | 371 | 36 | 308 | Glycosyl hydrolase family 76. Family of alpha-1,6-mannanases. |
| 224250 | YyaL | 1.17e-07 | 165 | 381 | 274 | 515 | Uncharacterized conserved protein YyaL, SSP411 family, contains thoiredoxin and six-hairpin glycosidase-like domains [General function prediction only]. |
| 227170 | COG4833 | 5.12e-07 | 158 | 325 | 87 | 260 | Predicted alpha-1,6-mannanase, GH76 family [Carbohydrate transport and metabolism]. |
| 224250 | YyaL | 6.94e-07 | 161 | 313 | 408 | 567 | Uncharacterized conserved protein YyaL, SSP411 family, contains thoiredoxin and six-hairpin glycosidase-like domains [General function prediction only]. |
| 400034 | Glyco_hydro_88 | 2.25e-06 | 115 | 267 | 29 | 178 | Glycosyl Hydrolase Family 88. Unsaturated glucuronyl hydrolase catalyzes the hydrolytic release of unsaturated glucuronic acids from oligosaccharides (EC:3.2.1.-) produced by the reactions of polysaccharide lyases. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 4.32e-232 | 5 | 434 | 4 | 444 | |
| 5.58e-135 | 77 | 432 | 176 | 527 | |
| 3.95e-131 | 77 | 432 | 182 | 533 | |
| 2.34e-109 | 76 | 434 | 52 | 415 | |
| 1.55e-101 | 73 | 433 | 207 | 572 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5.78e-15 | 126 | 312 | 63 | 267 | Glycoside hydrolase family 76 (mannosidase) Bt3792 from Bacteroides thetaiotaomicron VPI-5482 [Bacteroides thetaiotaomicron VPI-5482],4C1S_B Glycoside hydrolase family 76 (mannosidase) Bt3792 from Bacteroides thetaiotaomicron VPI-5482 [Bacteroides thetaiotaomicron VPI-5482] |
|
| 1.29e-13 | 61 | 312 | 124 | 391 | Crystal Structure of a family 76 glycoside hydrolase from a bovine Bacteroides thetaiotaomicron strain [Bacteroides thetaiotaomicron] |
|
| 3.28e-12 | 162 | 341 | 116 | 300 | Crystal structure of a putative glycosylhydrolase (BT_3782) from Bacteroides thetaiotaomicron VPI-5482 at 1.89 A resolution [Bacteroides thetaiotaomicron VPI-5482],4MU9_B Crystal structure of a putative glycosylhydrolase (BT_3782) from Bacteroides thetaiotaomicron VPI-5482 at 1.89 A resolution [Bacteroides thetaiotaomicron VPI-5482] |
|
| 8.71e-10 | 156 | 317 | 127 | 293 | Structure of the GH76A alpha-1,6-mannanase from Salegentibacter sp. HEL1_6 [Salegentibacter sp. Hel_I_6],6SHD_B Structure of the GH76A alpha-1,6-mannanase from Salegentibacter sp. HEL1_6 [Salegentibacter sp. Hel_I_6],6SHD_C Structure of the GH76A alpha-1,6-mannanase from Salegentibacter sp. HEL1_6 [Salegentibacter sp. Hel_I_6] |
|
| 5.38e-09 | 77 | 328 | 19 | 256 | Chain A, Alpha-1,6-mannanase [Niallia circulans],4BOJ_B Chain B, Alpha-1,6-mannanase [Niallia circulans],4BOJ_C Chain C, Alpha-1,6-mannanase [Niallia circulans] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7.11e-10 | 160 | 432 | 100 | 408 | Meiotically up-regulated gene 191 protein OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=mug191 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
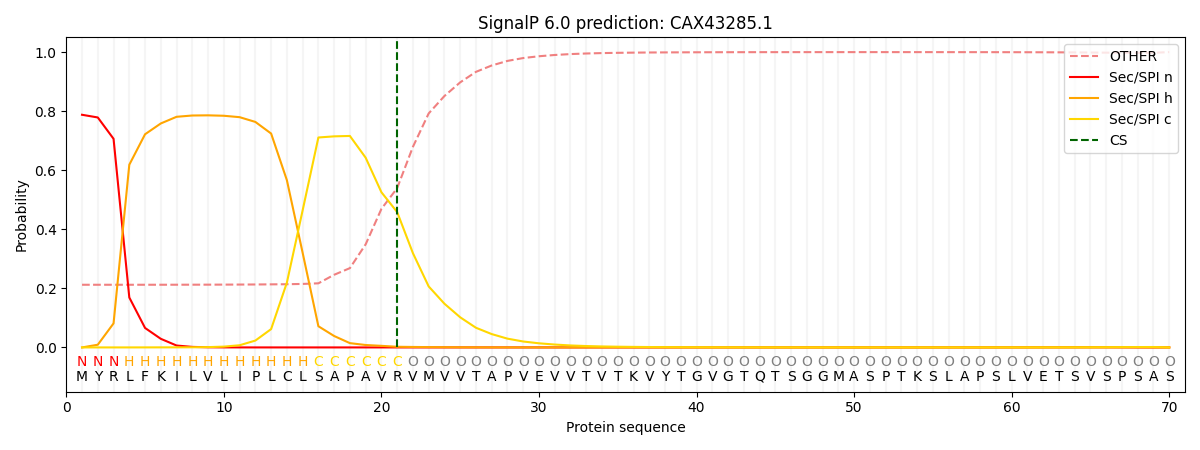
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.238133 | 0.761849 | CS pos: 21-22. Pr: 0.4583 |
