You are browsing environment: FUNGIDB
CAZyme Information: CAX40420.1
You are here: Home > Sequence: CAX40420.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Candida dubliniensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Saccharomycetes; ; Debaryomycetaceae; Candida; Candida dubliniensis | |||||||||||
| CAZyme ID | CAX40420.1 | |||||||||||
| CAZy Family | GH13 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.21:4 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH3 | 26 | 248 | 1.5e-61 | 0.9861111111111112 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 185053 | PRK15098 | 3.49e-51 | 29 | 806 | 101 | 758 | beta-glucosidase BglX. |
| 224389 | BglX | 5.90e-48 | 28 | 403 | 56 | 382 | Periplasmic beta-glucosidase and related glycosidases [Carbohydrate transport and metabolism]. |
| 396478 | Glyco_hydro_3_C | 1.45e-47 | 354 | 589 | 1 | 216 | Glycosyl hydrolase family 3 C-terminal domain. This domain is involved in catalysis and may be involved in binding beta-glucan. This domain is found associated with pfam00933. |
| 178629 | PLN03080 | 1.45e-33 | 23 | 772 | 72 | 742 | Probable beta-xylosidase; Provisional |
| 395747 | Glyco_hydro_3 | 1.08e-32 | 29 | 288 | 63 | 314 | Glycosyl hydrolase family 3 N terminal domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 815 | 110 | 924 | |
| 0.0 | 1 | 815 | 93 | 906 | |
| 0.0 | 1 | 815 | 93 | 906 | |
| 0.0 | 1 | 814 | 60 | 871 | |
| 0.0 | 1 | 814 | 60 | 871 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.57e-224 | 1 | 807 | 34 | 831 | Chain A, Beta-glucosidase [Thermochaetoides thermophila] |
|
| 3.53e-220 | 1 | 807 | 36 | 834 | Three-dimensional structures of two heavily N-glycosylated Aspergillus sp. Family GH3 beta-D-glucosidases [Aspergillus oryzae],5FJJ_B Three-dimensional structures of two heavily N-glycosylated Aspergillus sp. Family GH3 beta-D-glucosidases [Aspergillus oryzae],5FJJ_C Three-dimensional structures of two heavily N-glycosylated Aspergillus sp. Family GH3 beta-D-glucosidases [Aspergillus oryzae],5FJJ_D Three-dimensional structures of two heavily N-glycosylated Aspergillus sp. Family GH3 beta-D-glucosidases [Aspergillus oryzae] |
|
| 4.83e-220 | 1 | 807 | 35 | 833 | Crystal structure of beta-glucosidase 1 from Aspergillus aculeatus [Aspergillus aculeatus],4IIB_B Crystal structure of beta-glucosidase 1 from Aspergillus aculeatus [Aspergillus aculeatus],4IIC_A Crystal structure of beta-glucosidase 1 from Aspergillus aculeatus in complex with isofagomine [Aspergillus aculeatus],4IIC_B Crystal structure of beta-glucosidase 1 from Aspergillus aculeatus in complex with isofagomine [Aspergillus aculeatus],4IID_A Crystal structure of beta-glucosidase 1 from Aspergillus aculeatus in complex with 1-deoxynojirimycin [Aspergillus aculeatus],4IID_B Crystal structure of beta-glucosidase 1 from Aspergillus aculeatus in complex with 1-deoxynojirimycin [Aspergillus aculeatus],4IIE_A Crystal structure of beta-glucosidase 1 from Aspergillus aculeatus in complex with calystegine B(2) [Aspergillus aculeatus],4IIE_B Crystal structure of beta-glucosidase 1 from Aspergillus aculeatus in complex with calystegine B(2) [Aspergillus aculeatus],4IIF_A Crystal structure of beta-glucosidase 1 from Aspergillus aculeatus in complex with castanospermine [Aspergillus aculeatus],4IIF_B Crystal structure of beta-glucosidase 1 from Aspergillus aculeatus in complex with castanospermine [Aspergillus aculeatus],4IIG_A Crystal structure of beta-glucosidase 1 from Aspergillus aculeatus in complex with D-glucose [Aspergillus aculeatus],4IIG_B Crystal structure of beta-glucosidase 1 from Aspergillus aculeatus in complex with D-glucose [Aspergillus aculeatus],4IIH_A Crystal structure of beta-glucosidase 1 from Aspergillus aculeatus in complex with thiocellobiose [Aspergillus aculeatus],4IIH_B Crystal structure of beta-glucosidase 1 from Aspergillus aculeatus in complex with thiocellobiose [Aspergillus aculeatus] |
|
| 1.14e-219 | 1 | 807 | 50 | 852 | Structural studies of a Glycoside Hydrolase Family 3 beta-glucosidase from the Model Fungus Neurospora crassa [Neurospora crassa OR74A],5NBS_B Structural studies of a Glycoside Hydrolase Family 3 beta-glucosidase from the Model Fungus Neurospora crassa [Neurospora crassa OR74A] |
|
| 5.13e-218 | 1 | 807 | 54 | 847 | Chain A, Beta-glucosidase [Rasamsonia emersonii],5JU6_B Chain B, Beta-glucosidase [Rasamsonia emersonii],5JU6_C Chain C, Beta-glucosidase [Rasamsonia emersonii],5JU6_D Chain D, Beta-glucosidase [Rasamsonia emersonii] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6.37e-294 | 1 | 798 | 70 | 865 | Beta-glucosidase 2 OS=Saccharomycopsis fibuligera OX=4944 GN=BGL2 PE=3 SV=1 |
|
| 1.78e-287 | 1 | 798 | 68 | 861 | Beta-glucosidase 1 OS=Saccharomycopsis fibuligera OX=4944 GN=BGL1 PE=3 SV=1 |
|
| 2.48e-224 | 1 | 807 | 54 | 852 | Beta-glucosidase A OS=Aspergillus kawachii (strain NBRC 4308) OX=1033177 GN=bglA PE=1 SV=2 |
|
| 1.26e-221 | 1 | 807 | 54 | 852 | Probable beta-glucosidase A OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=bglA PE=3 SV=1 |
|
| 4.17e-220 | 1 | 811 | 55 | 857 | Probable beta-glucosidase A OS=Aspergillus terreus OX=33178 GN=bglA PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
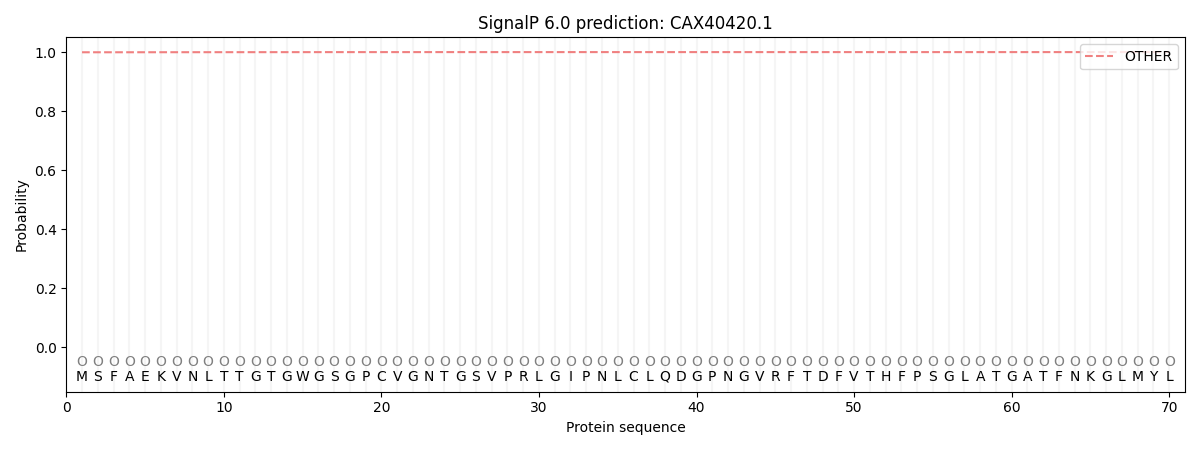
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.999543 | 0.000467 |
