You are browsing environment: FUNGIDB
CAZyme Information: CAGL0J11044g-T-p1
You are here: Home > Sequence: CAGL0J11044g-T-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | [Candida] glabrata | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Saccharomycetes; ; Debaryomycetaceae; Candida; [Candida] glabrata | |||||||||||
| CAZyme ID | CAGL0J11044g-T-p1 | |||||||||||
| CAZy Family | GT91 | |||||||||||
| CAZyme Description | Has domain(s) with predicted endoplasmic reticulum membrane localization | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 224839 | PMT1 | 1.48e-25 | 124 | 736 | 112 | 664 | Dolichyl-phosphate-mannose--protein O-mannosyl transferase [Posttranslational modification, protein turnover, chaperones]. |
| 197746 | MIR | 1.41e-06 | 383 | 436 | 4 | 54 | Domain in ryanodine and inositol trisphosphate receptors and protein O-mannosyltransferases. |
| 397103 | MIR | 1.69e-05 | 369 | 518 | 50 | 182 | MIR domain. The MIR (protein mannosyltransferase, IP3R and RyR) domain is a domain that may have a ligand transferase function. |
| 406576 | PMT_4TMC | 0.003 | 614 | 736 | 57 | 162 | C-terminal four TMM region of protein-O-mannosyltransferase. PMT_4TMC is the C-terminal four membrane-pass region of protein-O-mannosyltransferases and similar enzymes. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 736 | 1 | 736 | |
| 0.0 | 1 | 736 | 1 | 736 | |
| 0.0 | 1 | 736 | 1 | 736 | |
| 0.0 | 1 | 736 | 1 | 736 | |
| 0.0 | 1 | 736 | 1 | 736 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.18e-41 | 28 | 719 | 13 | 662 | Probable dolichyl-phosphate-mannose--protein mannosyltransferase 7 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=PMT7 PE=1 SV=1 |
|
| 6.04e-13 | 325 | 523 | 347 | 527 | Dolichyl-phosphate-mannose--protein mannosyltransferase 1 OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=PMT1 PE=2 SV=1 |
|
| 1.07e-08 | 311 | 523 | 295 | 494 | Dolichyl-phosphate-mannose--protein mannosyltransferase 5 OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=PMT5 PE=2 SV=1 |
|
| 5.42e-07 | 225 | 436 | 227 | 430 | Dolichyl-phosphate-mannose--protein mannosyltransferase 1 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=ogm1 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000062 | 0.000000 |
TMHMM Annotations download full data without filtering help
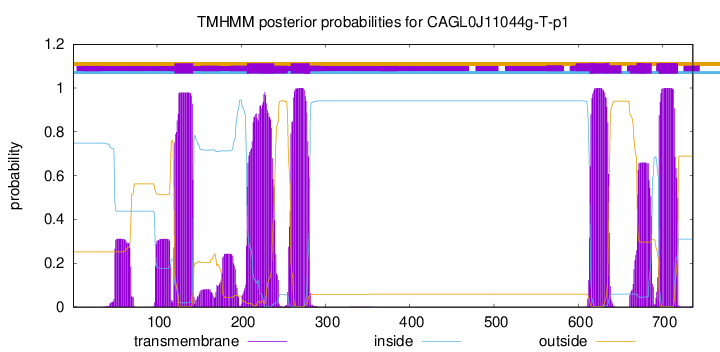
| Start | End |
|---|---|
| 121 | 143 |
| 207 | 239 |
| 259 | 281 |
| 614 | 636 |
| 670 | 687 |
| 696 | 718 |
