You are browsing environment: FUNGIDB
CAZyme Information: CAG86689.2
You are here: Home > Sequence: CAG86689.2
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Debaryomyces hansenii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Saccharomycetes; ; Debaryomycetaceae; Debaryomyces; Debaryomyces hansenii | |||||||||||
| CAZyme ID | CAG86689.2 | |||||||||||
| CAZy Family | GH18 | |||||||||||
| CAZyme Description | DEHA2D02068p [Source:UniProtKB/TrEMBL;Acc:Q6BTB3] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH76 | 138 | 399 | 4.1e-32 | 0.7402234636871509 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 397638 | Glyco_hydro_76 | 2.23e-21 | 162 | 388 | 33 | 284 | Glycosyl hydrolase family 76. Family of alpha-1,6-mannanases. |
| 224250 | YyaL | 5.18e-07 | 215 | 440 | 274 | 520 | Uncharacterized conserved protein YyaL, SSP411 family, contains thoiredoxin and six-hairpin glycosidase-like domains [General function prediction only]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 9.47e-127 | 123 | 493 | 208 | 576 | |
| 7.77e-123 | 124 | 493 | 210 | 577 | |
| 2.24e-122 | 124 | 493 | 51 | 418 | |
| 1.71e-105 | 124 | 488 | 87 | 442 | |
| 4.58e-105 | 126 | 491 | 182 | 536 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.66e-06 | 200 | 292 | 71 | 171 | Chain A, Alpha-1,6-mannanase [Niallia circulans] |
|
| 4.71e-06 | 200 | 292 | 74 | 174 | Chain A, Alpha-1,6-mannanase [Niallia circulans],4BOJ_B Chain B, Alpha-1,6-mannanase [Niallia circulans],4BOJ_C Chain C, Alpha-1,6-mannanase [Niallia circulans] |
|
| 4.97e-06 | 200 | 292 | 92 | 192 | Chain A, Alpha-1,6-mannanase [Niallia circulans],4D4A_B Chain B, Alpha-1,6-mannanase [Niallia circulans],4D4B_A Chain A, Alpha-1,6-mannanase [Niallia circulans],4D4B_B Chain B, Alpha-1,6-mannanase [Niallia circulans],4D4C_A Chain A, Alpha-1,6-mannanase [Niallia circulans],4D4C_B Chain B, Alpha-1,6-mannanase [Niallia circulans],4D4D_A Chain A, Alpha-1,6-mannanase [Niallia circulans],4D4D_B Chain B, Alpha-1,6-mannanase [Niallia circulans],5N0F_A Chain A, Alpha-1,6-mannanase [Niallia circulans],5N0F_B Chain B, Alpha-1,6-mannanase [Niallia circulans],6ZBX_A Chain A, Alpha-1,6-mannanase [Niallia circulans],6ZBX_B Chain B, Alpha-1,6-mannanase [Niallia circulans],7NL5_A Chain A, Alpha-1,6-mannanase [Niallia circulans] |
|
| 6.94e-06 | 200 | 292 | 105 | 205 | Chain A, Alpha-1,6-mannanase [Niallia circulans],5M77_B Chain B, Alpha-1,6-mannanase [Niallia circulans] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.92e-10 | 163 | 493 | 46 | 413 | Meiotically up-regulated gene 191 protein OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=mug191 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
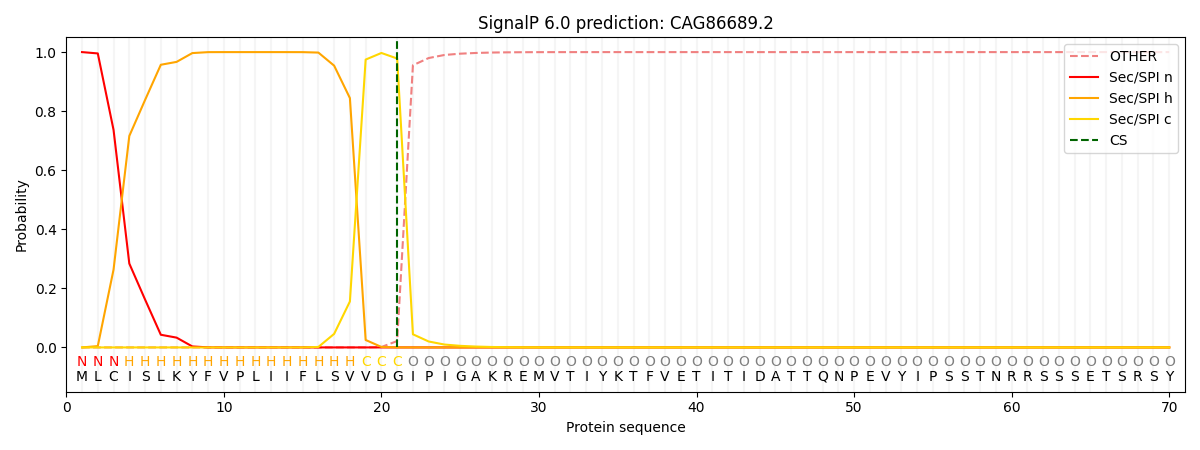
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000320 | 0.999671 | CS pos: 21-22. Pr: 0.9783 |
