You are browsing environment: FUNGIDB
CAZyme Information: CAG84319.2
You are here: Home > Sequence: CAG84319.2
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Debaryomyces hansenii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Saccharomycetes; ; Debaryomycetaceae; Debaryomyces; Debaryomyces hansenii | |||||||||||
| CAZyme ID | CAG84319.2 | |||||||||||
| CAZy Family | AA1 | |||||||||||
| CAZyme Description | DEHA2A00792p [Source:UniProtKB/TrEMBL;Acc:Q6BZJ7] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT91 | 140 | 628 | 9.3e-161 | 0.9955654101995566 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 403385 | DUF3589 | 0.0 | 139 | 628 | 1 | 485 | Protein of unknown function (DUF3589). This family of proteins is found in eukaryotes. Proteins in this family are typically between 541 and 717 amino acids in length. The function of this family is not known, |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 640 | 1 | 640 | |
| 7.11e-238 | 1 | 638 | 7 | 636 | |
| 6.59e-160 | 68 | 638 | 1 | 548 | |
| 5.50e-154 | 65 | 638 | 54 | 604 | |
| 1.00e-153 | 66 | 639 | 82 | 692 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7.51e-131 | 68 | 638 | 78 | 640 | Beta-mannosyltransferase 4 OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=BMT4 PE=2 SV=2 |
|
| 1.58e-126 | 116 | 633 | 94 | 647 | Beta-mannosyltransferase 8 OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=BMT8 PE=3 SV=1 |
|
| 8.92e-126 | 122 | 638 | 133 | 634 | Beta-mannosyltransferase 6 OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=BMT6 PE=2 SV=2 |
|
| 3.46e-115 | 62 | 634 | 50 | 601 | Beta-mannosyltransferase 5 OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=BMT5 PE=2 SV=1 |
|
| 2.40e-113 | 68 | 640 | 82 | 638 | Beta-mannosyltransferase 2 OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=RHD1 PE=2 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
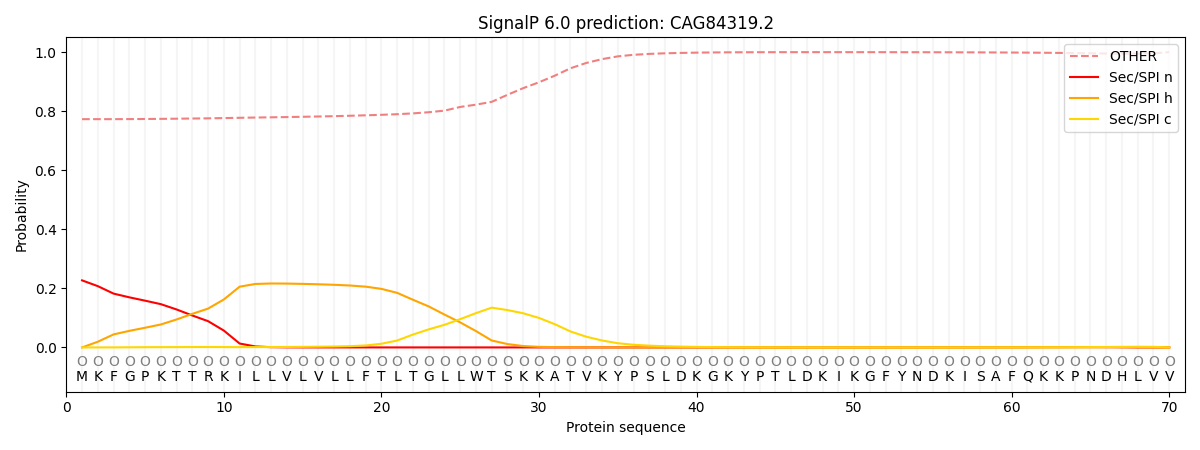
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.780804 | 0.219213 |

