You are browsing environment: FUNGIDB
CAZyme Information: C2_10150W_A-T-p1
You are here: Home > Sequence: C2_10150W_A-T-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Candida albicans | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Saccharomycetes; ; Debaryomycetaceae; Candida; Candida albicans | |||||||||||
| CAZyme ID | C2_10150W_A-T-p1 | |||||||||||
| CAZy Family | GH3 | |||||||||||
| CAZyme Description | Secreted protein, fluconazole-induced | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH76 | 66 | 310 | 2.9e-31 | 0.7122905027932961 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 397638 | Glyco_hydro_76 | 7.78e-15 | 99 | 307 | 59 | 294 | Glycosyl hydrolase family 76. Family of alpha-1,6-mannanases. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.84e-300 | 1 | 401 | 1 | 401 | |
| 1.03e-155 | 23 | 399 | 25 | 410 | |
| 8.47e-149 | 31 | 400 | 37 | 416 | |
| 1.22e-125 | 29 | 399 | 21 | 383 | |
| 8.74e-121 | 29 | 399 | 21 | 383 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6.05e-14 | 124 | 301 | 116 | 300 | Crystal structure of a putative glycosylhydrolase (BT_3782) from Bacteroides thetaiotaomicron VPI-5482 at 1.89 A resolution [Bacteroides thetaiotaomicron VPI-5482],4MU9_B Crystal structure of a putative glycosylhydrolase (BT_3782) from Bacteroides thetaiotaomicron VPI-5482 at 1.89 A resolution [Bacteroides thetaiotaomicron VPI-5482] |
|
| 9.38e-14 | 124 | 271 | 133 | 286 | Structure of the GH76A alpha-1,6-mannanase from Salegentibacter sp. HEL1_6 [Salegentibacter sp. Hel_I_6],6SHD_B Structure of the GH76A alpha-1,6-mannanase from Salegentibacter sp. HEL1_6 [Salegentibacter sp. Hel_I_6],6SHD_C Structure of the GH76A alpha-1,6-mannanase from Salegentibacter sp. HEL1_6 [Salegentibacter sp. Hel_I_6] |
|
| 3.02e-13 | 119 | 271 | 128 | 287 | Chain A, Alpha-1,6-endo-mannanase GH76A mutant [Salegentibacter sp. Hel_I_6] |
|
| 4.04e-13 | 136 | 271 | 146 | 287 | An inactive (D136A and D137A) variant of alpha-1,6-mannanase, GH76A of Salegentibacter sp. HEL1_6 in complex with alpha-1,6-mannotetrose [Salegentibacter sp. Hel_I_6] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.27e-06 | 40 | 254 | 8 | 248 | Meiotically up-regulated gene 191 protein OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=mug191 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
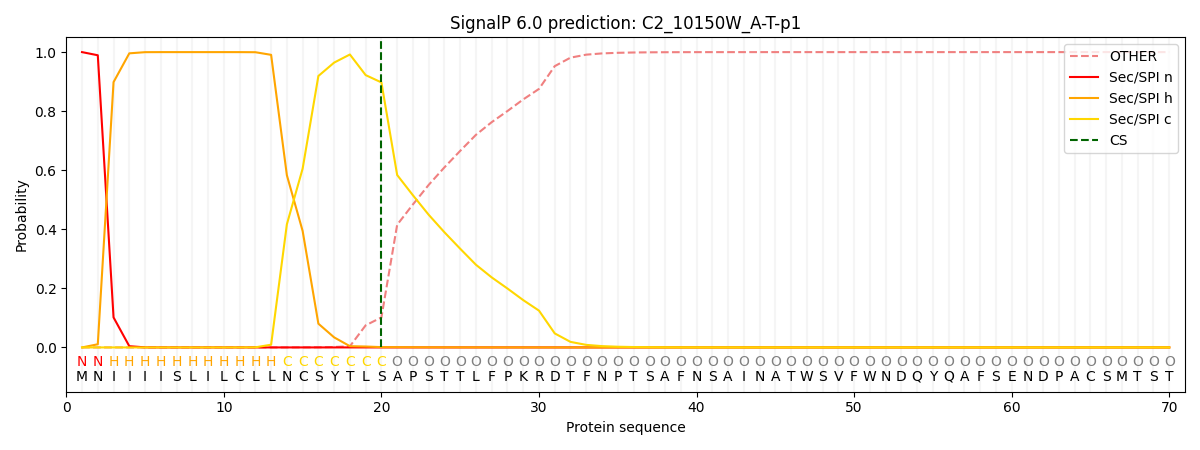
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000215 | 0.999750 | CS pos: 20-21. Pr: 0.8973 |
