You are browsing environment: FUNGIDB
CAZyme Information: C1_02420C_B-T-p1
You are here: Home > Sequence: C1_02420C_B-T-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Candida albicans | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Saccharomycetes; ; Debaryomycetaceae; Candida; Candida albicans | |||||||||||
| CAZyme ID | C1_02420C_B-T-p1 | |||||||||||
| CAZy Family | AA2 | |||||||||||
| CAZyme Description | Essential beta-1,3-glucan synthase subunit, gsc1 allele determines resistance/sensitivity to echinocandins, 16 predicted membrane-spanning regions, mRNA abundance declines after yeast-to-hypha transition, Spider biofilm induced | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 2.4.1.34:56 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT48 | 810 | 1594 | 0 | 0.9891745602165088 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 396784 | Glucan_synthase | 0.0 | 810 | 1643 | 1 | 819 | 1,3-beta-glucan synthase component. This family consists of various 1,3-beta-glucan synthase components including Gls1, Gls2 and Gls3 from yeast. 1,3-beta-glucan synthase EC:2.4.1.34 also known as callose synthase catalyzes the formation of a beta-1,3-glucan polymer that is a major component of the fungal cell wall. The reaction catalyzed is:- UDP-glucose + {(1,3)-beta-D-glucosyl}(N) <=> UDP + {(1,3)-beta-D-glucosyl}(N+1). |
| 405046 | FKS1_dom1 | 8.28e-47 | 302 | 411 | 1 | 111 | 1,3-beta-glucan synthase subunit FKS1, domain-1. The FKS1_dom1 domain is likely to be the 'Class I' region just N-terminal to the first set of transmembrane helices that is involved in 1,3-beta-glucan synthesis itself. This family is found on proteins with family Glucan_synthase, pfam02364. |
| 312941 | Med15 | 1.47e-06 | 13 | 143 | 201 | 333 | ARC105 or Med15 subunit of Mediator complex non-fungal. The approx. 70 residue Med15 domain of the ARC-Mediator co-activator is a three-helix bundle with marked similarity to the KIX domain. The sterol regulatory element binding protein (SREBP) family of transcription activators use the ARC105 subunit to activate target genes in the regulation of cholesterol and fatty acid homeostasis. In addition, Med15 is a critical transducer of gene activation signals that control early metazoan development. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 1897 | 1 | 1897 | |
| 0.0 | 1 | 1897 | 1 | 1897 | |
| 0.0 | 1 | 1897 | 1 | 1897 | |
| 0.0 | 1 | 1897 | 1 | 1897 | |
| 0.0 | 1 | 1897 | 1 | 1897 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 0.0 | 33 | 1879 | 36 | 1894 | 1,3-beta-glucan synthase component GSC2 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=GSC2 PE=1 SV=2 |
|
| 0.0 | 81 | 1866 | 96 | 1872 | 1,3-beta-glucan synthase component FKS1 OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=fksA PE=3 SV=1 |
|
| 0.0 | 144 | 1873 | 11 | 1770 | 1,3-beta-glucan synthase component FKS3 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=FKS3 PE=1 SV=1 |
|
| 0.0 | 81 | 1879 | 86 | 1875 | 1,3-beta-glucan synthase component FKS1 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=FKS1 PE=1 SV=2 |
|
| 0.0 | 99 | 1832 | 95 | 1781 | 1,3-beta-glucan synthase component FKS1 OS=Cryptococcus neoformans var. grubii serotype A (strain H99 / ATCC 208821 / CBS 10515 / FGSC 9487) OX=235443 GN=FKS1 PE=3 SV=3 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000073 | 0.000001 |
TMHMM Annotations download full data without filtering help
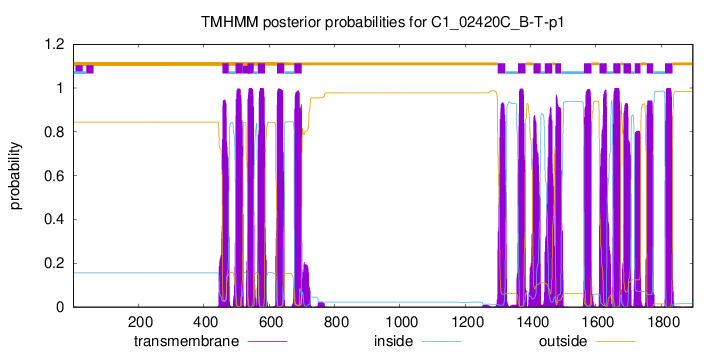
| Start | End |
|---|---|
| 457 | 476 |
| 497 | 519 |
| 534 | 552 |
| 565 | 587 |
| 624 | 646 |
| 677 | 699 |
| 1300 | 1322 |
| 1362 | 1384 |
| 1409 | 1431 |
| 1444 | 1466 |
| 1476 | 1493 |
| 1564 | 1586 |
| 1611 | 1633 |
| 1653 | 1675 |
| 1685 | 1707 |
| 1719 | 1736 |
| 1756 | 1775 |
| 1812 | 1834 |
