You are browsing environment: FUNGIDB
CAZyme Information: Bcin11g04000.1:RNA-p1
You are here: Home > Sequence: Bcin11g04000.1:RNA-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
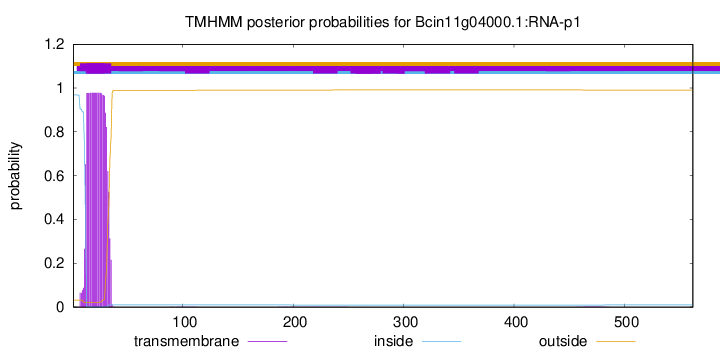
TMHMM annotations
Basic Information help
| Species | Botrytis cinerea | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Leotiomycetes; ; Sclerotiniaceae; Botrytis; Botrytis cinerea | |||||||||||
| CAZyme ID | Bcin11g04000.1:RNA-p1 | |||||||||||
| CAZy Family | GT8 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.28:5 |
|---|
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 224471 | ATH1 | 1.11e-28 | 95 | 561 | 32 | 459 | Trehalose and maltose hydrolase (possible phosphorylase) [Carbohydrate transport and metabolism]. |
| 397616 | Glyco_hydro_65N | 9.65e-25 | 95 | 354 | 19 | 240 | Glycosyl hydrolase family 65, N-terminal domain. This family of glycosyl hydrolases contains vacuolar acid trehalase and maltose phosphorylase.Maltose phosphorylase (MP) is a dimeric enzyme that catalyzes the conversion of maltose and inorganic phosphate into beta-D-glucose-1-phosphate and glucose. This domain is believed to be essential for catalytic activity although its precise function remains unknown. |
| 237517 | PRK13807 | 3.53e-15 | 156 | 498 | 74 | 395 | maltose phosphorylase; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 562 | 1 | 562 | |
| 0.0 | 10 | 562 | 1 | 553 | |
| 3.26e-268 | 35 | 562 | 22 | 551 | |
| 1.41e-129 | 60 | 558 | 138 | 630 | |
| 9.58e-122 | 69 | 558 | 31 | 516 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5.19e-09 | 336 | 560 | 234 | 434 | Chain A, Candidate alpha glycoside phosphorylase Glycoside hydrolase family 65 [Flavobacterium johnsoniae UW101],7FE3_B Chain B, Candidate alpha glycoside phosphorylase Glycoside hydrolase family 65 [Flavobacterium johnsoniae UW101],7FE3_C Chain C, Candidate alpha glycoside phosphorylase Glycoside hydrolase family 65 [Flavobacterium johnsoniae UW101],7FE4_A Chain A, Candidate alpha glycoside phosphorylase Glycoside hydrolase family 65 [Flavobacterium johnsoniae UW101],7FE4_B Chain B, Candidate alpha glycoside phosphorylase Glycoside hydrolase family 65 [Flavobacterium johnsoniae UW101],7FE4_C Chain C, Candidate alpha glycoside phosphorylase Glycoside hydrolase family 65 [Flavobacterium johnsoniae UW101] |
|
| 2.37e-06 | 330 | 560 | 222 | 439 | Chain A, Glycoside hydrolase family 65 central catalytic [[Bacillus] selenitireducens MLS10],4KTR_B Chain B, Glycoside hydrolase family 65 central catalytic [[Bacillus] selenitireducens MLS10],4KTR_C Chain C, Glycoside hydrolase family 65 central catalytic [[Bacillus] selenitireducens MLS10],4KTR_D Chain D, Glycoside hydrolase family 65 central catalytic [[Bacillus] selenitireducens MLS10],4KTR_E Chain E, Glycoside hydrolase family 65 central catalytic [[Bacillus] selenitireducens MLS10],4KTR_F Chain F, Glycoside hydrolase family 65 central catalytic [[Bacillus] selenitireducens MLS10],4KTR_G Chain G, Glycoside hydrolase family 65 central catalytic [[Bacillus] selenitireducens MLS10],4KTR_H Chain H, Glycoside hydrolase family 65 central catalytic [[Bacillus] selenitireducens MLS10] |
|
| 5.42e-06 | 330 | 560 | 222 | 439 | Chain A, Glycoside hydrolase family 65 central catalytic [[Bacillus] selenitireducens MLS10],4KTP_B Chain B, Glycoside hydrolase family 65 central catalytic [[Bacillus] selenitireducens MLS10] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.67e-94 | 71 | 560 | 58 | 550 | Acid trehalase OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=treA PE=3 SV=1 |
|
| 2.65e-84 | 71 | 562 | 61 | 554 | Cell wall acid trehalase ARB_03719 OS=Arthroderma benhamiae (strain ATCC MYA-4681 / CBS 112371) OX=663331 GN=ARB_03719 PE=1 SV=1 |
|
| 9.83e-57 | 75 | 541 | 107 | 550 | Cell wall acid trehalase ATC1 OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=ATC1 PE=1 SV=2 |
|
| 1.51e-44 | 33 | 541 | 133 | 619 | Periplasmic/secreted acid trehalase ATH1 OS=Candida glabrata OX=5478 GN=ATH1 PE=1 SV=1 |
|
| 4.19e-42 | 61 | 541 | 115 | 586 | Periplasmic acid trehalase ATC1 OS=Saccharomyces cerevisiae (strain CEN.PK113-7D) OX=889517 GN=CENPK1137D_1727 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.083628 | 0.916371 | CS pos: 32-33. Pr: 0.8663 |