You are browsing environment: FUNGIDB
CAZyme Information: B9J08_003123-t37_1-p1
You are here: Home > Sequence: B9J08_003123-t37_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | [Candida] auris | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Saccharomycetes; ; Debaryomycetaceae; Candida; [Candida] auris | |||||||||||
| CAZyme ID | B9J08_003123-t37_1-p1 | |||||||||||
| CAZy Family | GT22 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 2.4.1.131:20 | 2.4.1.-:1 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT15 | 99 | 378 | 1.1e-117 | 0.9926739926739927 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 227353 | KTR1 | 2.75e-152 | 99 | 433 | 80 | 398 | Mannosyltransferase [Carbohydrate transport and metabolism]. |
| 396385 | Glyco_transf_15 | 6.50e-117 | 97 | 379 | 40 | 313 | Glycolipid 2-alpha-mannosyltransferase. This is a family of alpha-1,2 mannosyl-transferases involved in N-linked and O-linked glycosylation of proteins. Some of the enzymes in this family have been shown to be involved in O- and N-linked glycan modifications in the Golgi. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 438 | 1 | 438 | |
| 0.0 | 1 | 439 | 1 | 439 | |
| 0.0 | 1 | 439 | 1 | 439 | |
| 1.10e-303 | 53 | 439 | 1 | 387 | |
| 1.08e-256 | 1 | 418 | 1 | 418 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.41e-104 | 98 | 404 | 25 | 324 | Crystal structure of yeast alpha1,2-mannosyltransferase Kre2p/Mnt1p [Saccharomyces cerevisiae],1S4N_B Crystal structure of yeast alpha1,2-mannosyltransferase Kre2p/Mnt1p [Saccharomyces cerevisiae],1S4O_A Crystal structure of yeast alpha1,2-mannosyltransferase Kre2p/Mnt1p: binary complex with GDP/Mn [Saccharomyces cerevisiae],1S4O_B Crystal structure of yeast alpha1,2-mannosyltransferase Kre2p/Mnt1p: binary complex with GDP/Mn [Saccharomyces cerevisiae],1S4P_A Crystal structure of yeast alpha1,2-mannosyltransferase Kre2p/Mnt1p: ternary complex with GDP/Mn and methyl-alpha-mannoside acceptor [Saccharomyces cerevisiae],1S4P_B Crystal structure of yeast alpha1,2-mannosyltransferase Kre2p/Mnt1p: ternary complex with GDP/Mn and methyl-alpha-mannoside acceptor [Saccharomyces cerevisiae] |
|
| 2.82e-91 | 86 | 386 | 53 | 352 | Crystal Structure of Core-mannan synthase A (CmsA/Ktr4) from Aspergillus fumigatus, apo form [Aspergillus fumigatus A1163],7BOO_B Crystal Structure of Core-mannan synthase A (CmsA/Ktr4) from Aspergillus fumigatus, apo form [Aspergillus fumigatus A1163],7BOO_C Crystal Structure of Core-mannan synthase A (CmsA/Ktr4) from Aspergillus fumigatus, apo form [Aspergillus fumigatus A1163],7BOO_D Crystal Structure of Core-mannan synthase A (CmsA/Ktr4) from Aspergillus fumigatus, apo form [Aspergillus fumigatus A1163],7BOO_E Crystal Structure of Core-mannan synthase A (CmsA/Ktr4) from Aspergillus fumigatus, apo form [Aspergillus fumigatus A1163],7BOO_F Crystal Structure of Core-mannan synthase A (CmsA/Ktr4) from Aspergillus fumigatus, apo form [Aspergillus fumigatus A1163],7BOP_A Crystal Structure of Core-mannan synthase A (CmsA/Ktr4) from Aspergillus fumigatus, Mn/GDP-form [Aspergillus fumigatus A1163],7BOP_B Crystal Structure of Core-mannan synthase A (CmsA/Ktr4) from Aspergillus fumigatus, Mn/GDP-form [Aspergillus fumigatus A1163],7BOP_C Crystal Structure of Core-mannan synthase A (CmsA/Ktr4) from Aspergillus fumigatus, Mn/GDP-form [Aspergillus fumigatus A1163],7BOP_D Crystal Structure of Core-mannan synthase A (CmsA/Ktr4) from Aspergillus fumigatus, Mn/GDP-form [Aspergillus fumigatus A1163],7BOP_E Crystal Structure of Core-mannan synthase A (CmsA/Ktr4) from Aspergillus fumigatus, Mn/GDP-form [Aspergillus fumigatus A1163],7BOP_F Crystal Structure of Core-mannan synthase A (CmsA/Ktr4) from Aspergillus fumigatus, Mn/GDP-form [Aspergillus fumigatus A1163] |
|
| 1.19e-80 | 98 | 431 | 101 | 431 | X-ray structure of the mannosyltransferase Ktr4p from S. cerevisiae in complex with GDP [Saccharomyces cerevisiae S288C],5A07_B X-ray structure of the mannosyltransferase Ktr4p from S. cerevisiae in complex with GDP [Saccharomyces cerevisiae S288C],5A08_A X-ray structure of the mannosyltransferase Ktr4p from S. cerevisiae [Saccharomyces cerevisiae S288C],5A08_B X-ray structure of the mannosyltransferase Ktr4p from S. cerevisiae [Saccharomyces cerevisiae S288C] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 9.53e-132 | 59 | 431 | 49 | 421 | Probable mannosyltransferase KTR2 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=KTR2 PE=3 SV=1 |
|
| 9.69e-130 | 99 | 433 | 90 | 423 | Probable mannosyltransferase YUR1 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=YUR1 PE=1 SV=1 |
|
| 5.85e-109 | 71 | 422 | 42 | 377 | Alpha-1,2 mannosyltransferase KTR1 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=KTR1 PE=1 SV=1 |
|
| 1.02e-108 | 77 | 421 | 45 | 372 | O-glycoside alpha-1,2-mannosyltransferase homolog 3 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=omh3 PE=3 SV=1 |
|
| 1.10e-107 | 99 | 437 | 108 | 430 | Glycolipid 2-alpha-mannosyltransferase 1 OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=MNT1 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
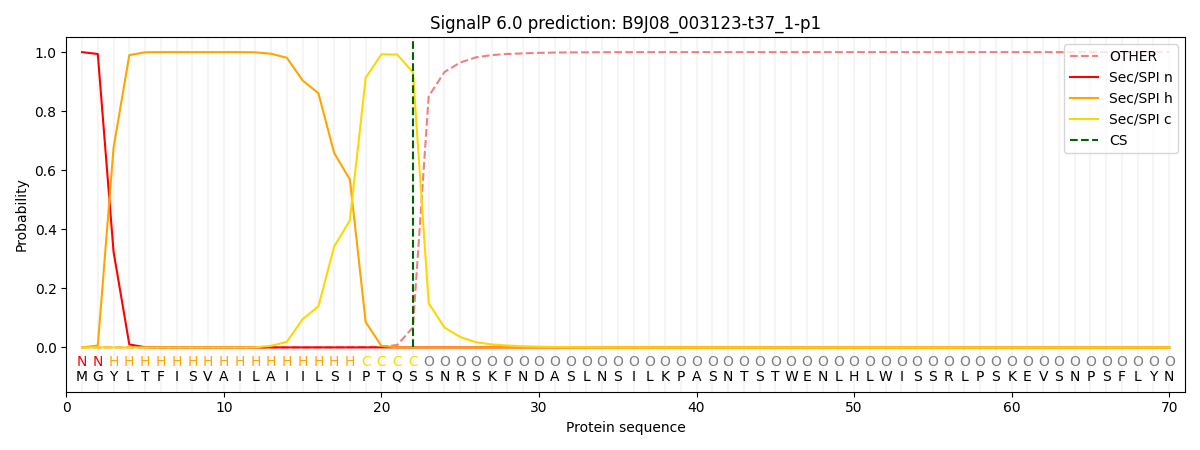
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000719 | 0.999255 | CS pos: 22-23. Pr: 0.9319 |
