You are browsing environment: FUNGIDB
CAZyme Information: An14g07390-T-p1
You are here: Home > Sequence: An14g07390-T-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aspergillus niger | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus niger | |||||||||||
| CAZyme ID | An14g07390-T-p1 | |||||||||||
| CAZy Family | GT1 | |||||||||||
| CAZyme Description | Has domain(s) with predicted hydrolase activity, hydrolyzing O-glycosyl compounds activity and role in carbohydrate metabolic process | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.8:39 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH11 | 33 | 208 | 2.9e-60 | 0.9774011299435028 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 395367 | Glyco_hydro_11 | 1.17e-66 | 30 | 206 | 1 | 175 | Glycosyl hydrolases family 11. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.01e-92 | 1 | 211 | 1 | 211 | |
| 1.01e-92 | 1 | 211 | 1 | 211 | |
| 1.01e-92 | 1 | 211 | 1 | 211 | |
| 8.25e-92 | 1 | 211 | 1 | 211 | |
| 8.25e-92 | 1 | 211 | 1 | 211 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.22e-88 | 1 | 211 | 1 | 211 | Crystal structure of Aspergillus niger GH11 endoxylanase XynA in complex with xylobiose epoxide activity based probe [Aspergillus niger] |
|
| 8.91e-72 | 28 | 211 | 1 | 184 | Endo-1,4-Beta-Xylanase C [Aspergillus awamori],1T6G_C Chain C, Endo-1,4-beta-xylanase I [Aspergillus niger],1T6G_D Chain D, Endo-1,4-beta-xylanase I [Aspergillus niger] |
|
| 5.29e-71 | 28 | 211 | 2 | 185 | Xylanase C from Aspergillus kawachii F131W mutant [Aspergillus luchuensis] |
|
| 2.07e-70 | 28 | 211 | 1 | 184 | Structure Of Endo-1,4-Beta-Xylanase C [Aspergillus niger],1UKR_B Structure Of Endo-1,4-Beta-Xylanase C [Aspergillus niger],1UKR_C Structure Of Endo-1,4-Beta-Xylanase C [Aspergillus niger],1UKR_D Structure Of Endo-1,4-Beta-Xylanase C [Aspergillus niger] |
|
| 2.94e-70 | 28 | 211 | 1 | 184 | Chain A, Endo-1,4-beta-xylanase I [Aspergillus niger] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.79e-93 | 1 | 211 | 1 | 211 | Probable endo-1,4-beta-xylanase 5 OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=XYN5 PE=3 SV=1 |
|
| 3.77e-88 | 1 | 211 | 1 | 211 | Endo-1,4-beta-xylanase A OS=Aspergillus niger OX=5061 GN=xynA PE=1 SV=1 |
|
| 2.17e-87 | 1 | 211 | 1 | 211 | Endo-1,4-beta-xylanase 3 OS=Aspergillus kawachii (strain NBRC 4308) OX=1033177 GN=xynC PE=1 SV=2 |
|
| 2.17e-87 | 1 | 211 | 1 | 211 | Endo-1,4-beta-xylanase 5 OS=Aspergillus niger OX=5061 GN=XYN5 PE=1 SV=1 |
|
| 2.17e-87 | 1 | 211 | 1 | 211 | Endo-1,4-beta-xylanase A OS=Aspergillus awamori OX=105351 GN=xynA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
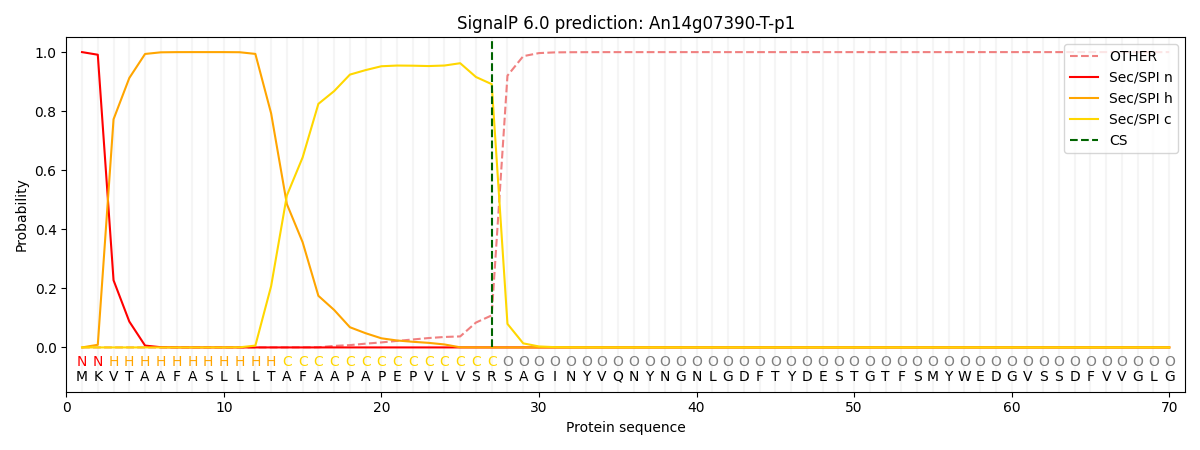
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000225 | 0.999745 | CS pos: 27-28. Pr: 0.8915 |
