You are browsing environment: FUNGIDB
CAZyme Information: An14g01390-T-p1
Basic Information
help
| Species |
Aspergillus niger
|
| Lineage |
Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus niger
|
| CAZyme ID |
An14g01390-T-p1
|
| CAZy Family |
GH76 |
| CAZyme Description |
Ortholog of Aspergillus tubingensis : Asptu1_0132292, Aspergillus kawachii : Aspka1_0177360, Aspergillus acidus : Aspfo1_0048298 and Aspergillus niger ATCC 1015 : 41574-mRNA
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 684 |
|
76513.58 |
4.4526 |
|
| Genome Property |
| Genome Version/Assembly ID |
Genes |
Strain NCBI Taxon ID |
Non Protein Coding Genes |
Protein Coding Genes |
| FungiDB-61_AnigerCBS513-88 |
14403 |
425011 |
346 |
14057
|
|
| Gene Location |
No EC number prediction in An14g01390-T-p1.
| Family |
Start |
End |
Evalue |
family coverage |
| PL27 |
62 |
674 |
8e-223 |
0.9950819672131147 |
An14g01390-T-p1 has no CDD domain.
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
| 0.0 |
1 |
684 |
1 |
684 |
| 0.0 |
1 |
683 |
1 |
683 |
| 0.0 |
1 |
683 |
1 |
683 |
| 0.0 |
1 |
683 |
1 |
683 |
| 3.87e-196 |
26 |
675 |
7 |
662 |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
| 1.76e-58 |
66 |
681 |
69 |
694 |
Polysaccharide Lyase BACCELL_00875 [Bacteroides cellulosilyticus DSM 14838],5NO8_B Polysaccharide Lyase BACCELL_00875 [Bacteroides cellulosilyticus DSM 14838] |
| 9.99e-56 |
66 |
681 |
69 |
694 |
Polysaccharide Lyase BACCELL_00875 [Bacteroides cellulosilyticus DSM 14838],5NOK_B Polysaccharide Lyase BACCELL_00875 [Bacteroides cellulosilyticus DSM 14838] |
An14g01390-T-p1 has no Swissprot hit.
This protein is predicted as SP
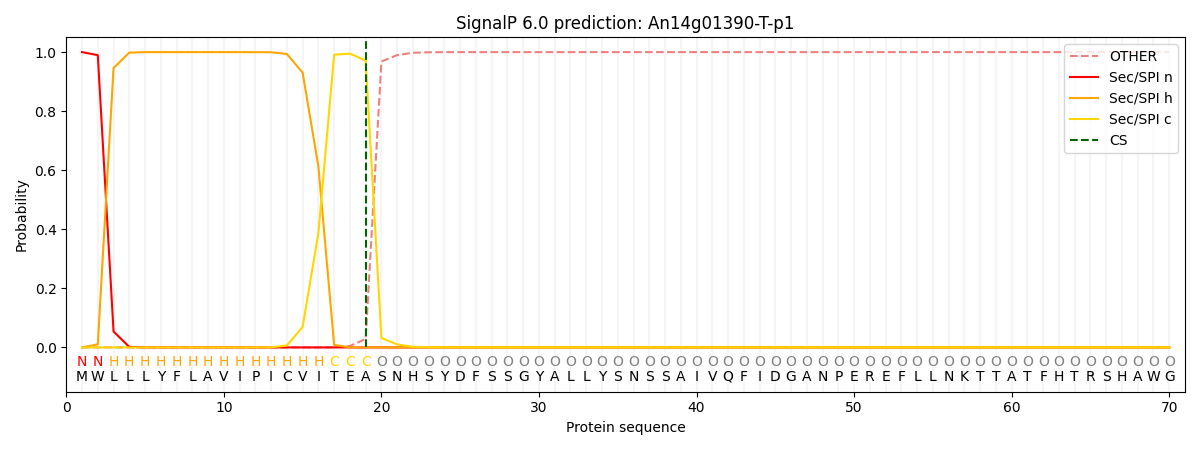
| Other |
SP_Sec_SPI |
CS Position |
| 0.000177 |
0.999793 |
CS pos: 19-20. Pr: 0.9715 |
There is no transmembrane helices in An14g01390-T-p1.

