You are browsing environment: FUNGIDB
CAZyme Information: An08g04330-T-p1
You are here: Home > Sequence: An08g04330-T-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aspergillus niger | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus niger | |||||||||||
| CAZyme ID | An08g04330-T-p1 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | Ortholog of Aspergillus tubingensis : Asptu1_0037715, Aspergillus brasiliensis : Aspbr1_0053866, Aspergillus kawachii : Aspka1_0178640 and Aspergillus acidus : Aspfo1_0129898 | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.109:1 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH114 | 34 | 224 | 1.1e-71 | 0.9947368421052631 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 397552 | Glyco_hydro_114 | 2.06e-96 | 33 | 263 | 1 | 220 | Glycoside-hydrolase family GH114. This family is recognized as a glycosyl-hydrolase family, number 114. It is endo-alpha-1,4-polygalactosaminidase, a rare enzyme. It is proposed to be TIM-barrel, the most common structure amongst the catalytic domains of glycosyl-hydrolases. |
| 226386 | COG3868 | 3.79e-27 | 52 | 224 | 83 | 266 | Predicted glycosyl hydrolase, GH114 family [Carbohydrate transport and metabolism]. |
| 225219 | COG2342 | 1.89e-06 | 61 | 258 | 60 | 281 | Endo alpha-1,4 polygalactosaminidase, GH114 family (was erroneously annotated as Cys-tRNA synthetase) [Carbohydrate transport and metabolism]. |
| 273582 | TIGR01370 | 1.64e-05 | 61 | 233 | 84 | 282 | extracellular protein. Original assignment of this protein family as cysteinyl-tRNA synthetase is controversial, supported by but challenged by and by subsequent discovery of the actual mechanism for synthesizing Cys-tRNA in species where a direct Cys--tRNA ligase was not found. Lingering legacy annotations of members of this family probably should be removed. Evidence against the role includes a signal peptide. This family as been renamed "extracellular protein" to facilitate correction. Members of this family occur in Deinococcus radiodurans (bacterial) and Methanococcus jannaschii (archaeal). A number of homologous but more distantly related proteins are annotated as alpha-1,4 polygalactosaminidases. The function remains unknown. [Unknown function, General] |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.87e-206 | 1 | 270 | 1 | 270 | |
| 3.93e-179 | 1 | 270 | 1 | 270 | |
| 6.33e-179 | 1 | 270 | 14 | 283 | |
| 2.28e-102 | 24 | 269 | 71 | 316 | |
| 3.29e-95 | 3 | 269 | 3 | 276 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.93e-92 | 21 | 269 | 25 | 277 | Crystal Structure of Aspergillus fumigatus Ega3 [Aspergillus fumigatus Af293] |
|
| 5.10e-92 | 21 | 269 | 42 | 294 | Crystal Structure of Aspergillus fumigatus Ega3 complex with galactosamine [Aspergillus fumigatus Af293] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
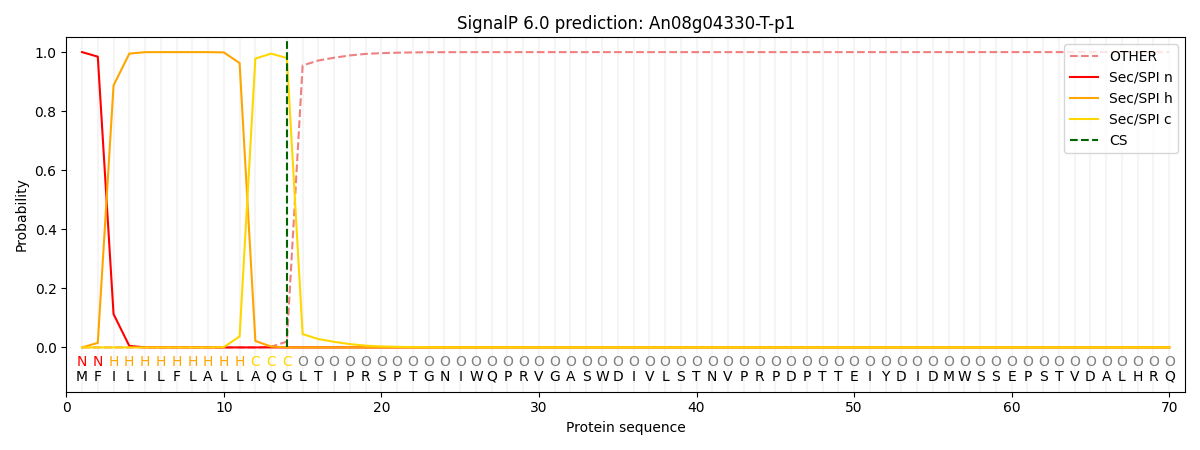
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000230 | 0.999733 | CS pos: 14-15. Pr: 0.9801 |
