You are browsing environment: FUNGIDB
CAZyme Information: An06g01140-T-p1
You are here: Home > Sequence: An06g01140-T-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aspergillus niger | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus niger | |||||||||||
| CAZyme ID | An06g01140-T-p1 | |||||||||||
| CAZy Family | GH135 | |||||||||||
| CAZyme Description | Has domain(s) with predicted transferase activity, transferring hexosyl groups activity and role in metabolic process | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT1 | 149 | 452 | 1.4e-31 | 0.680628272251309 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 224732 | YjiC | 1.57e-38 | 28 | 454 | 13 | 397 | UDP:flavonoid glycosyltransferase YjiC, YdhE family [Carbohydrate transport and metabolism]. |
| 340817 | GT1_Gtf-like | 1.67e-28 | 19 | 441 | 3 | 391 | UDP-glycosyltransferases and similar proteins. This family includes the Gtfs, a group of homologous glycosyltransferases involved in the final stages of the biosynthesis of antibiotics vancomycin and related chloroeremomycin. Gtfs transfer sugar moieties from an activated NDP-sugar donor to the oxidatively cross-linked heptapeptide core of vancomycin group antibiotics. The core structure is important for the bioactivity of the antibiotics. |
| 273616 | MGT | 9.34e-24 | 28 | 441 | 7 | 375 | glycosyltransferase, MGT family. This model describes the MGT (macroside glycosyltransferase) subfamily of the UDP-glucuronosyltransferase family. Members include a number of glucosyl transferases for macrolide antibiotic inactivation, but also include transferases of glucose-related sugars for macrolide antibiotic production. [Cellular processes, Toxin production and resistance] |
| 278624 | UDPGT | 4.91e-08 | 244 | 429 | 237 | 412 | UDP-glucoronosyl and UDP-glucosyl transferase. |
| 223071 | egt | 0.002 | 252 | 431 | 264 | 438 | ecdysteroid UDP-glucosyltransferase; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 456 | 1 | 456 | |
| 3.84e-284 | 1 | 456 | 1 | 455 | |
| 3.84e-284 | 1 | 456 | 1 | 455 | |
| 2.66e-123 | 10 | 454 | 114 | 556 | |
| 6.41e-123 | 16 | 453 | 25 | 459 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.89e-11 | 28 | 438 | 32 | 389 | Crystal Structure of CalG1, Calicheamicin Glycostyltransferase, TDP bound form [Micromonospora echinospora],3OTH_A Crystal Structure of CalG1, Calicheamicin Glycostyltransferase, TDP and calicheamicin alpha3I bound form [Micromonospora echinospora],3OTH_B Crystal Structure of CalG1, Calicheamicin Glycostyltransferase, TDP and calicheamicin alpha3I bound form [Micromonospora echinospora] |
|
| 3.15e-11 | 283 | 438 | 222 | 376 | Crystal Structure Analysis of the Glycotransferase of kitacinnamycin [Kitasatospora],6J31_B Crystal Structure Analysis of the Glycotransferase of kitacinnamycin [Kitasatospora],6J31_C Crystal Structure Analysis of the Glycotransferase of kitacinnamycin [Kitasatospora],6J31_D Crystal Structure Analysis of the Glycotransferase of kitacinnamycin [Kitasatospora] |
|
| 3.19e-11 | 283 | 438 | 225 | 379 | Crystal Structure Analysis of the Glycotransferase of kitacinnamycin [Kitasatospora],6J32_B Crystal Structure Analysis of the Glycotransferase of kitacinnamycin [Kitasatospora],6J32_C Crystal Structure Analysis of the Glycotransferase of kitacinnamycin [Kitasatospora],6J32_D Crystal Structure Analysis of the Glycotransferase of kitacinnamycin [Kitasatospora] |
|
| 3.62e-11 | 288 | 448 | 269 | 424 | Structural Characterization of UDP-glycosyltransferase from Tetranychus Urticae [Tetranychus urticae] |
|
| 2.47e-08 | 339 | 430 | 317 | 408 | Structure of the glycosyltransferase EryCIII from the erythromycin biosynthetic pathway, in complex with its activating partner, EryCII [Saccharopolyspora erythraea NRRL 2338] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.38e-103 | 28 | 453 | 30 | 455 | UDP-glucosyltransferase 1 OS=Beauveria bassiana (strain ARSEF 2860) OX=655819 GN=BBA_08686 PE=2 SV=1 |
|
| 5.89e-70 | 16 | 454 | 5 | 429 | UDP-glucosyltransferase B1 OS=Starmerella bombicola OX=75736 GN=ugtB1 PE=1 SV=1 |
|
| 1.31e-69 | 16 | 454 | 7 | 433 | UDP-glucosyltransferase A1 OS=Starmerella bombicola OX=75736 GN=ugtA1 PE=1 SV=1 |
|
| 2.39e-53 | 19 | 445 | 5 | 410 | 4'-demethylrebeccamycin synthase OS=Lentzea aerocolonigenes OX=68170 GN=rebG PE=1 SV=1 |
|
| 4.84e-08 | 289 | 430 | 225 | 364 | Uncharacterized UDP-glucosyltransferase YdhE OS=Bacillus subtilis (strain 168) OX=224308 GN=ydhE PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
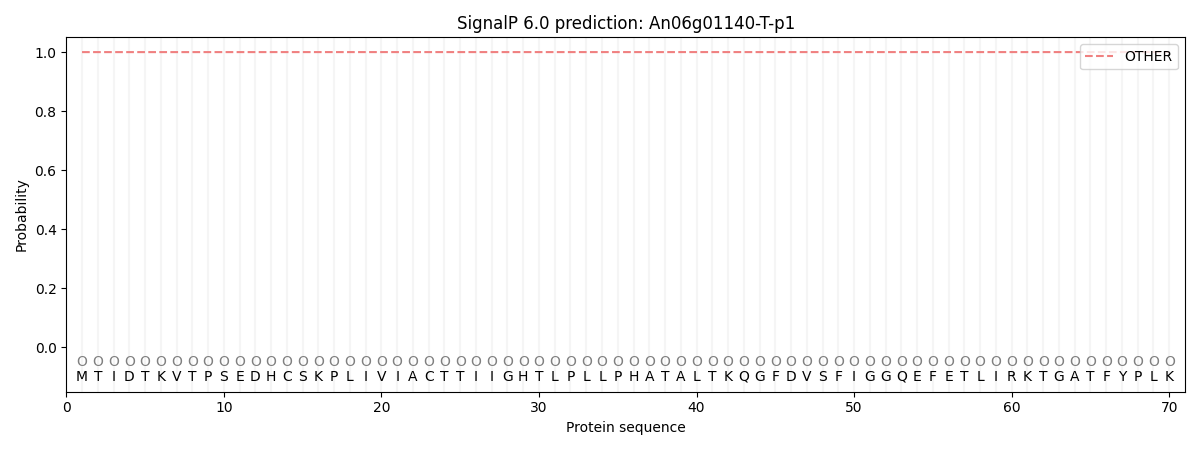
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000019 | 0.000016 |
