You are browsing environment: FUNGIDB
CAZyme Information: An02g02980-T-p1
You are here: Home > Sequence: An02g02980-T-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aspergillus niger | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus niger | |||||||||||
| CAZyme ID | An02g02980-T-p1 | |||||||||||
| CAZy Family | AA7 | |||||||||||
| CAZyme Description | Ortholog(s) have endoplasmic reticulum localization | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT59 | 34 | 429 | 3.7e-131 | 0.8985148514851485 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 398537 | DIE2_ALG10 | 8.24e-121 | 35 | 431 | 1 | 349 | DIE2/ALG10 family. The ALG10 protein from Saccharomyces cerevisiae encodes the alpha-1,2 glucosyltransferase of the endoplasmic reticulum. This protein has been characterized in rat as potassium channel regulator 1. |
| 411490 | pneumo_PspA | 2.70e-04 | 440 | 505 | 354 | 432 | pneumococcal surface protein A. The pneumococcal surface protein proteins, found in Streptococcus pneumoniae, are repetitive, with patterns of localized high sequence identity across pairs of proteins given different specific names that recombination may be presumed. This protein, PspA, has an N-terminal region that lacks a cross-wall-targeting YSIRK type extended signal peptide, in contrast to the closely related choline-binding protein CbpA which has a similar C-terminus but a YSIRK-containing region at the N-terminus. |
| 225629 | FtsN | 0.001 | 428 | 478 | 129 | 182 | Cell division protein FtsN [Cell cycle control, cell division, chromosome partitioning]. |
| 236940 | PRK11633 | 0.001 | 429 | 477 | 87 | 135 | cell division protein DedD; Provisional |
| 236940 | PRK11633 | 0.002 | 428 | 477 | 91 | 141 | cell division protein DedD; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 566 | 1 | 566 | |
| 0.0 | 1 | 566 | 1 | 608 | |
| 0.0 | 1 | 566 | 1 | 608 | |
| 5.72e-287 | 3 | 566 | 4 | 608 | |
| 1.63e-286 | 3 | 566 | 4 | 608 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.25e-268 | 4 | 566 | 5 | 614 | Dol-P-Glc:Glc(2)Man(9)GlcNAc(2)-PP-Dol alpha-1,2-glucosyltransferase OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=alg10 PE=3 SV=1 |
|
| 7.72e-254 | 1 | 566 | 1 | 608 | Dol-P-Glc:Glc(2)Man(9)GlcNAc(2)-PP-Dol alpha-1,2-glucosyltransferase OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=alg10 PE=3 SV=1 |
|
| 9.13e-79 | 27 | 566 | 60 | 660 | Dol-P-Glc:Glc(2)Man(9)GlcNAc(2)-PP-Dol alpha-1,2-glucosyltransferase OS=Magnaporthe oryzae (strain 70-15 / ATCC MYA-4617 / FGSC 8958) OX=242507 GN=ALG10 PE=3 SV=1 |
|
| 9.51e-73 | 2 | 566 | 43 | 722 | Dol-P-Glc:Glc(2)Man(9)GlcNAc(2)-PP-Dol alpha-1,2-glucosyltransferase OS=Gibberella zeae (strain ATCC MYA-4620 / CBS 123657 / FGSC 9075 / NRRL 31084 / PH-1) OX=229533 GN=ALG10 PE=3 SV=1 |
|
| 7.61e-67 | 27 | 421 | 61 | 530 | Dol-P-Glc:Glc(2)Man(9)GlcNAc(2)-PP-Dol alpha-1,2-glucosyltransferase OS=Neurospora crassa (strain ATCC 24698 / 74-OR23-1A / CBS 708.71 / DSM 1257 / FGSC 987) OX=367110 GN=alg-10 PE=3 SV=3 |
SignalP and Lipop Annotations help
This protein is predicted as SP
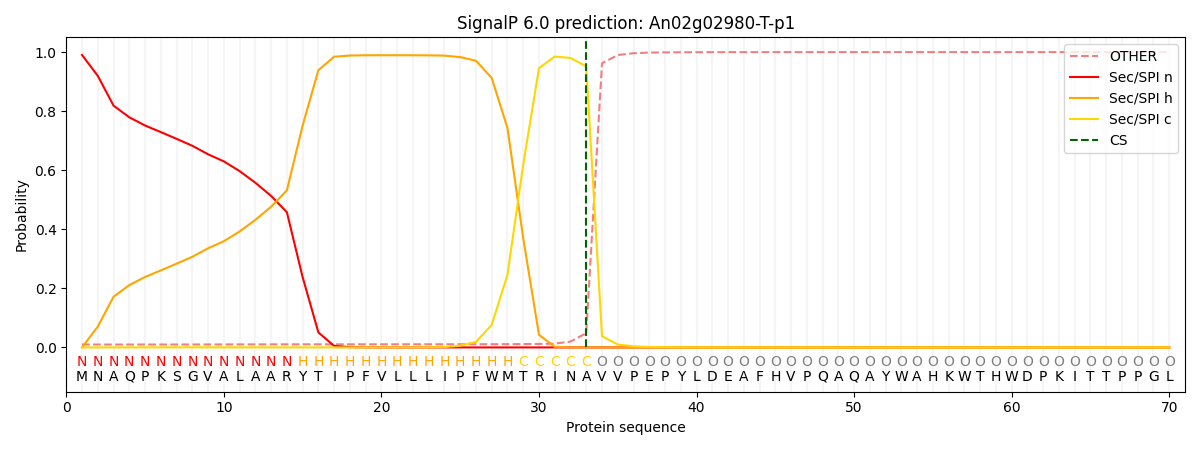
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.011601 | 0.988370 | CS pos: 33-34. Pr: 0.9518 |
TMHMM Annotations download full data without filtering help
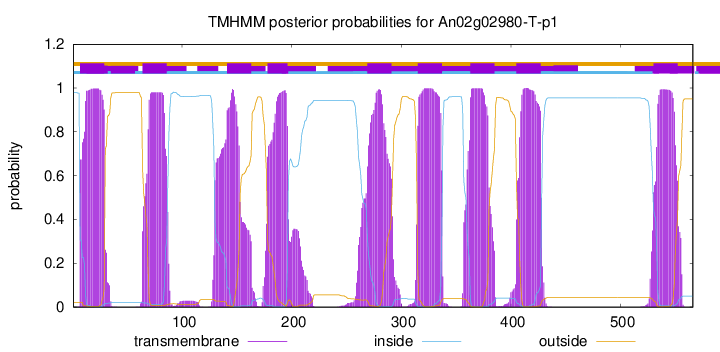
| Start | End |
|---|---|
| 7 | 29 |
| 64 | 86 |
| 141 | 163 |
| 178 | 196 |
| 269 | 291 |
| 315 | 337 |
| 363 | 385 |
| 405 | 427 |
| 530 | 552 |
