You are browsing environment: FUNGIDB
CAZyme Information: Afu7g02380-T-p1
You are here: Home > Sequence: Afu7g02380-T-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aspergillus fumigatus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus fumigatus | |||||||||||
| CAZyme ID | Afu7g02380-T-p1 | |||||||||||
| CAZy Family | GT2 | |||||||||||
| CAZyme Description | Ortholog of Aspergillus fumigatus A1163 : AFUB_088950 | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE1 | 49 | 174 | 9.7e-19 | 0.5814977973568282 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 226040 | LpqC | 4.28e-20 | 44 | 195 | 42 | 191 | Poly(3-hydroxybutyrate) depolymerase [Secondary metabolites biosynthesis, transport and catabolism]. |
| 273828 | esterase_phb | 5.06e-09 | 51 | 158 | 1 | 114 | esterase, PHB depolymerase family. This model describes a subfamily among lipases of the ab-hydrolase family. This subfamily includes bacterial depolymerases for poly(3-hydroxybutyrate) (PHB) and related polyhydroxyalkanoates (PHA), as well as acetyl xylan esterases, feruloyl esterases, and others from fungi. [Fatty acid and phospholipid metabolism, Degradation] |
| 402228 | Esterase_phd | 1.83e-04 | 66 | 211 | 17 | 168 | Esterase PHB depolymerase. This family of proteins include acetyl xylan esterases (AXE), feruloyl esterases (FAE), and poly(3-hydroxybutyrate) (PHB) depolymerases. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.02e-21 | 18 | 188 | 231 | 388 | |
| 6.90e-21 | 15 | 188 | 223 | 383 | |
| 1.80e-20 | 20 | 188 | 232 | 387 | |
| 3.40e-20 | 18 | 188 | 233 | 390 | |
| 3.56e-19 | 12 | 166 | 37 | 190 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.15e-12 | 49 | 159 | 292 | 406 | Feruloyl esterase B OS=Piromyces equi OX=99929 GN=ESTA PE=2 SV=1 |
|
| 7.62e-12 | 20 | 167 | 26 | 165 | Probable feruloyl esterase C OS=Aspergillus clavatus (strain ATCC 1007 / CBS 513.65 / DSM 816 / NCTC 3887 / NRRL 1 / QM 1276 / 107) OX=344612 GN=faeC-2 PE=3 SV=1 |
|
| 3.65e-11 | 13 | 167 | 19 | 165 | Probable feruloyl esterase C OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=faeC PE=3 SV=1 |
|
| 3.65e-11 | 13 | 167 | 19 | 165 | Probable feruloyl esterase C OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=faeC PE=3 SV=1 |
|
| 9.16e-11 | 3 | 159 | 7 | 154 | Probable feruloyl esterase C OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=faeC PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
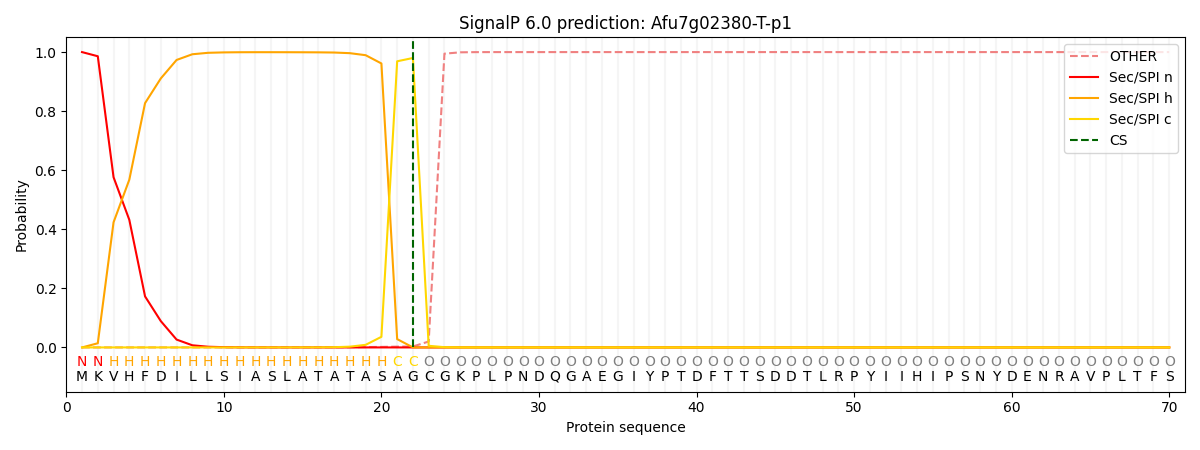
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000006 | 1.000036 | CS pos: 22-23. Pr: 0.9800 |
