You are browsing environment: FUNGIDB
CAZyme Information: Afu3g03610-T-p1
You are here: Home > Sequence: Afu3g03610-T-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
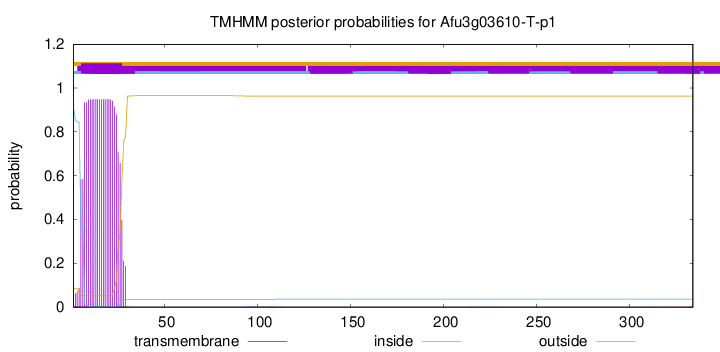
TMHMM annotations
Basic Information help
| Species | Aspergillus fumigatus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus fumigatus | |||||||||||
| CAZyme ID | Afu3g03610-T-p1 | |||||||||||
| CAZy Family | GH16 | |||||||||||
| CAZyme Description | Has domain(s) with predicted cellulase activity, hydrolase activity, hydrolyzing O-glycosyl compounds activity and role in polysaccharide catabolic process | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH12 | 182 | 332 | 7.1e-25 | 0.8782051282051282 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 396303 | Glyco_hydro_12 | 2.49e-10 | 85 | 333 | 3 | 206 | Glycosyl hydrolase family 12. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.84e-117 | 1 | 334 | 1 | 312 | |
| 1.84e-117 | 1 | 334 | 1 | 312 | |
| 1.84e-117 | 1 | 334 | 1 | 312 | |
| 1.84e-117 | 1 | 334 | 1 | 312 | |
| 4.25e-106 | 1 | 334 | 1 | 343 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.45e-13 | 82 | 331 | 19 | 226 | The X-ray Crystal Structure of the Trichoderma harzianum Endoglucanase 3 from family GH12 [Trichoderma harzianum],4H7M_B The X-ray Crystal Structure of the Trichoderma harzianum Endoglucanase 3 from family GH12 [Trichoderma harzianum] |
|
| 5.68e-11 | 85 | 317 | 16 | 203 | Chain A, GH12 beta-1, 4-endoglucanase [Aspergillus fischeri] |
|
| 6.43e-10 | 85 | 317 | 15 | 202 | Chain A, GH12 beta-1, 4-endoglucanase [Aspergillus fischeri] |
|
| 2.09e-07 | 85 | 317 | 15 | 207 | The structure of Aspergillus niger endoglucanase-palladium complex [Aspergillus niger],1KS5_A Structure of Aspergillus niger endoglucanase [Aspergillus niger] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.30e-09 | 85 | 319 | 36 | 227 | Xyloglucan-specific endo-beta-1,4-glucanase A OS=Aspergillus niger OX=5061 GN=xgeA PE=1 SV=1 |
|
| 1.30e-09 | 85 | 319 | 36 | 227 | Probable xyloglucan-specific endo-beta-1,4-glucanase A OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=xgeA PE=3 SV=1 |
|
| 3.46e-07 | 81 | 317 | 46 | 247 | Endoglucanase cel12B OS=Magnaporthe oryzae (strain 70-15 / ATCC MYA-4617 / FGSC 8958) OX=242507 GN=cel12B PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
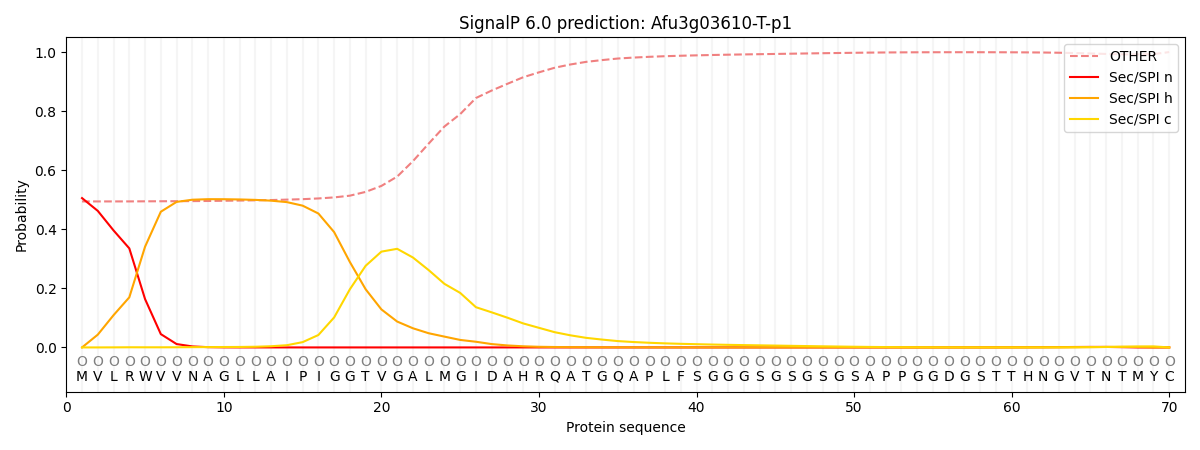
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.514603 | 0.485393 |