You are browsing environment: FUNGIDB
CAZyme Information: Afu1g00980-T-p1
You are here: Home > Sequence: Afu1g00980-T-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aspergillus fumigatus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus fumigatus | |||||||||||
| CAZyme ID | Afu1g00980-T-p1 | |||||||||||
| CAZy Family | AA11 | |||||||||||
| CAZyme Description | Monocarboxylate transporter, putative | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA7 | 57 | 296 | 1.5e-44 | 0.5021834061135371 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 223354 | GlcD | 1.05e-20 | 60 | 262 | 25 | 225 | FAD/FMN-containing dehydrogenase [Energy production and conversion]. |
| 396238 | FAD_binding_4 | 2.97e-17 | 72 | 212 | 5 | 139 | FAD binding domain. This family consists of various enzymes that use FAD as a co-factor, most of the enzymes are similar to oxygen oxidoreductase. One of the enzymes Vanillyl-alcohol oxidase (VAO) has a solved structure, the alignment includes the FAD binding site, called the PP-loop, between residues 99-110. The FAD molecule is covalently bound in the known structure, however the residue that links to the FAD is not in the alignment. VAO catalyzes the oxidation of a wide variety of substrates, ranging form aromatic amines to 4-alkylphenols. Other members of this family include D-lactate dehydrogenase, this enzyme catalyzes the conversion of D-lactate to pyruvate using FAD as a co-factor; mitomycin radical oxidase, this enzyme oxidizes the reduced form of mitomycins and is involved in mitomycin resistance. This family includes MurB an UDP-N-acetylenolpyruvoylglucosamine reductase enzyme EC:1.1.1.158. This enzyme is involved in the biosynthesis of peptidoglycan. |
| 273751 | FAD_lactone_ox | 1.71e-06 | 121 | 240 | 68 | 178 | sugar 1,4-lactone oxidases. This model represents a family of at least two different sugar 1,4 lactone oxidases, both involved in synthesizing ascorbic acid or a derivative. These include L-gulonolactone oxidase (EC 1.1.3.8) from rat and D-arabinono-1,4-lactone oxidase (EC 1.1.3.37) from Saccharomyces cerevisiae. Members are proposed to have the cofactor FAD covalently bound at a site specified by Prosite motif PS00862; OX2_COVAL_FAD; 1. |
| 369658 | BBE | 5.45e-06 | 451 | 493 | 3 | 44 | Berberine and berberine like. This domain is found in the berberine bridge and berberine bridge- like enzymes which are involved in the biosynthesis of numerous isoquinoline alkaloids. They catalyze the transformation of the N-methyl group of (S)-reticuline into the C-8 berberine bridge carbon of (S)-scoulerine. |
| 130737 | GLDHase | 5.42e-04 | 125 | 242 | 119 | 227 | galactonolactone dehydrogenase. This model represents L-Galactono-gamma-lactone dehydrogenase (EC 1.3.2.3). This enzyme catalyzes the final step in ascorbic acid biosynthesis in higher plants. This protein is homologous to ascorbic acid biosynthesis enzymes of other species: L-gulono-gamma-lactone oxidase in rat and L-galactono-gamma-lactone oxidase in yeast. All three covalently bind the cofactor FAD. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.23e-19 | 55 | 342 | 43 | 300 | |
| 2.27e-19 | 6 | 493 | 3 | 492 | |
| 2.27e-19 | 6 | 493 | 3 | 492 | |
| 1.84e-17 | 57 | 488 | 45 | 484 | |
| 6.07e-17 | 69 | 487 | 214 | 636 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.80e-22 | 47 | 495 | 27 | 470 | Physcomitrella patens BBE-like 1 variant D396N [Physcomitrium patens],6EO5_B Physcomitrella patens BBE-like 1 variant D396N [Physcomitrium patens] |
|
| 5.10e-22 | 47 | 495 | 27 | 470 | Physcomitrella patens BBE-like 1 wild-type [Physcomitrium patens],6EO4_B Physcomitrella patens BBE-like 1 wild-type [Physcomitrium patens] |
|
| 7.85e-21 | 6 | 493 | 3 | 492 | Xylooligosaccharide oxidase from Myceliophthora thermophila C1 [Thermothelomyces thermophilus ATCC 42464],5L6F_A Xylooligosaccharide oxidase from Myceliophthora thermophila C1 in complex with Xylobiose [Thermothelomyces thermophilus ATCC 42464],5L6G_A Xylooligosaccharide oxidase from Myceliophthora thermophila C1 in complex with Xylose [Thermothelomyces thermophilus ATCC 42464] |
|
| 1.88e-17 | 78 | 488 | 68 | 483 | Chain AAA, Chitooligosaccharide oxidase [Fusarium graminearum PH-1] |
|
| 2.98e-17 | 26 | 496 | 2 | 462 | The crystal structure of EncM T139V mutant [Streptomyces maritimus],6FYD_B The crystal structure of EncM T139V mutant [Streptomyces maritimus],6FYD_C The crystal structure of EncM T139V mutant [Streptomyces maritimus],6FYD_D The crystal structure of EncM T139V mutant [Streptomyces maritimus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 0.0 | 1 | 496 | 1 | 496 | FAD-linked oxidoreductase AFUA_1G00980 OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=AFUA_1G00980 PE=2 SV=1 |
|
| 3.70e-76 | 33 | 493 | 36 | 502 | FAD-linked oxidoreductase OXR1 OS=Magnaporthe oryzae (strain 70-15 / ATCC MYA-4617 / FGSC 8958) OX=242507 GN=OXR1 PE=1 SV=1 |
|
| 1.68e-70 | 39 | 496 | 45 | 498 | FAD-linked oxidoreductase srdI OS=Neurospora crassa (strain ATCC 24698 / 74-OR23-1A / CBS 708.71 / DSM 1257 / FGSC 987) OX=367110 GN=srdI PE=2 SV=1 |
|
| 1.34e-68 | 33 | 455 | 25 | 411 | FAD-linked oxidoreductase azaL OS=Aspergillus niger (strain ATCC 1015 / CBS 113.46 / FGSC A1144 / LSHB Ac4 / NCTC 3858a / NRRL 328 / USDA 3528.7) OX=380704 GN=azaL PE=2 SV=2 |
|
| 1.13e-62 | 40 | 493 | 1 | 450 | FAD-linked oxidoreductase virF (Fragment) OS=Hypocrea virens (strain Gv29-8 / FGSC 10586) OX=413071 GN=virF PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
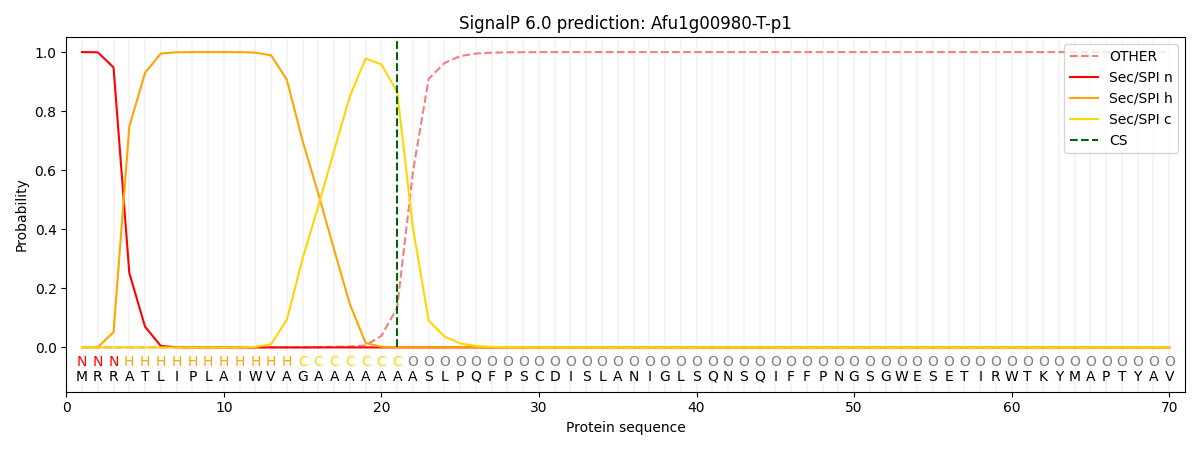
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000754 | 0.999250 | CS pos: 21-22. Pr: 0.8652 |
