You are browsing environment: FUNGIDB
CAZyme Information: AWU74113.1
You are here: Home > Sequence: AWU74113.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Pichia kudriavzevii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Saccharomycetes; ; Pichiaceae; Pichia; Pichia kudriavzevii | |||||||||||
| CAZyme ID | AWU74113.1 | |||||||||||
| CAZy Family | GT8 | |||||||||||
| CAZyme Description | Chitinase [Source:UniProtKB/TrEMBL;Acc:A0A2U9QYL1] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 119360 | GH18_PF-ChiA-like | 3.47e-41 | 3 | 259 | 3 | 292 | PF-ChiA is an uncharacterized chitinase found in the hyperthermophilic archaeon Pyrococcus furiosus with a glycosyl hydrolase family 18 (GH18) catalytic domain as well as a cellulose-binding domain. Members of this domain family are found not only in archaea but also in eukaryotes and prokaryotes. PF-ChiA exhibits hydrolytic activity toward both colloidal and crystalline (beta/alpha) chitins at high temperature. |
| 395573 | Glyco_hydro_18 | 0.002 | 33 | 237 | 39 | 307 | Glycosyl hydrolases family 18. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 4.06e-199 | 1 | 265 | 1 | 265 | |
| 1.38e-38 | 4 | 238 | 5 | 259 | |
| 2.99e-21 | 44 | 254 | 83 | 317 | |
| 4.02e-21 | 44 | 254 | 81 | 315 | |
| 5.41e-21 | 44 | 254 | 79 | 313 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.81e-13 | 53 | 243 | 69 | 269 | Crystal structure of catalytic domain of hyperthermophilic chitinase from Pyrococcus furiosus [Pyrococcus furiosus],2DSK_B Crystal structure of catalytic domain of hyperthermophilic chitinase from Pyrococcus furiosus [Pyrococcus furiosus] |
|
| 7.07e-13 | 53 | 243 | 69 | 269 | Crystal structures of catalytic site mutants of active domain 2 of thermostable chitinase from Pyrococcus furiosus complexed with chito-oligosaccharides [Pyrococcus furiosus],3A4W_B Crystal structures of catalytic site mutants of active domain 2 of thermostable chitinase from Pyrococcus furiosus complexed with chito-oligosaccharides [Pyrococcus furiosus] |
|
| 3.28e-12 | 53 | 243 | 69 | 269 | Crystal structures of catalytic site mutants of active domain 2 of thermostable chitinase from Pyrococcus furiosus complexed with chito-oligosaccharides [Pyrococcus furiosus],3A4X_B Crystal structures of catalytic site mutants of active domain 2 of thermostable chitinase from Pyrococcus furiosus complexed with chito-oligosaccharides [Pyrococcus furiosus],3AFB_A Crystal structures of catalytic site mutants of active domain 2 of chitinase from Pyrococcus furiosus [Pyrococcus furiosus],3AFB_B Crystal structures of catalytic site mutants of active domain 2 of chitinase from Pyrococcus furiosus [Pyrococcus furiosus] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as OTHER
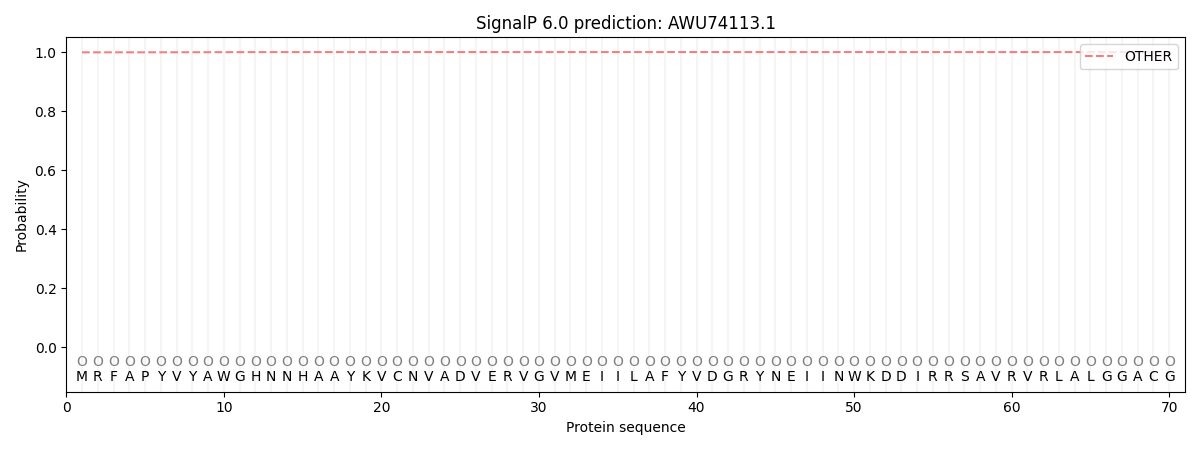
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.999323 | 0.000733 |
