You are browsing environment: FUNGIDB
CAZyme Information: ATY66787.1
You are here: Home > Sequence: ATY66787.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
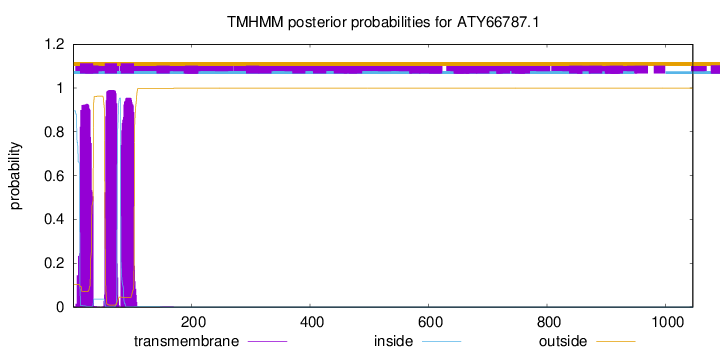
TMHMM annotations
Basic Information help
| Species | Cordyceps militaris | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Cordycipitaceae; Cordyceps; Cordyceps militaris | |||||||||||
| CAZyme ID | ATY66787.1 | |||||||||||
| CAZy Family | GT31 | |||||||||||
| CAZyme Description | beta-mannosidase mndA | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.25:4 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 131 | 880 | 1e-62 | 0.6529255319148937 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 225789 | LacZ | 3.77e-42 | 132 | 850 | 12 | 635 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| 407625 | Ig_mannosidase | 1.53e-17 | 965 | 1042 | 1 | 78 | Ig-fold domain. This Ig-like fold domain is found in mannosidase enzymes. |
| 407658 | Mannosidase_ig | 7.20e-08 | 859 | 915 | 2 | 53 | Mannosidase Ig/CBM-like domain. This domain corresponds to domain 4 in the structure of Bacteroides thetaiotaomicron beta-mannosidase, BtMan2A. This domain has an Ig-like fold. |
| 395572 | Glyco_hydro_2 | 1.44e-04 | 361 | 456 | 22 | 106 | Glycosyl hydrolases family 2. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 1046 | 1 | 1046 | |
| 0.0 | 132 | 1043 | 30 | 939 | |
| 0.0 | 113 | 1043 | 16 | 939 | |
| 0.0 | 115 | 1043 | 18 | 939 | |
| 0.0 | 132 | 1043 | 30 | 939 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 0.0 | 113 | 1043 | 16 | 939 | Structure of Fungal beta-mannosidase from Glycoside Hydrolase Family 2 of Trichoderma harzianum [Trichoderma harzianum],4UOJ_A Chain A, BETA-MANNOSIDASE GH2 [Trichoderma harzianum],4UOJ_B Chain B, BETA-MANNOSIDASE GH2 [Trichoderma harzianum] |
|
| 7.22e-102 | 134 | 842 | 13 | 675 | mouse beta-mannosidase (MANBA) [Mus musculus],6DDU_A mouse beta-mannosidase bound to beta-D-mannose (MANBA) [Mus musculus] |
|
| 8.26e-72 | 132 | 1021 | 14 | 833 | Crystal structure of the GH2 exo-beta-mannanase from Xanthomonas axonopodis pv. citri in complex with mannose [Xanthomonas citri pv. citri str. 306],6BYE_B Crystal structure of the GH2 exo-beta-mannanase from Xanthomonas axonopodis pv. citri in complex with mannose [Xanthomonas citri pv. citri str. 306] |
|
| 8.38e-72 | 132 | 1021 | 14 | 833 | Crystal structure of the GH2 exo-beta-mannanase from Xanthomonas axonopodis pv. citri [Xanthomonas citri pv. citri str. 306] |
|
| 5.09e-71 | 132 | 1021 | 12 | 831 | Crystal structure of the acid-base mutant (E477A) of the GH2 exo-beta-mannanase from Xanthomonas axonopodis pv. citri [Xanthomonas citri pv. citri str. 306],6BYI_B Crystal structure of the acid-base mutant (E477A) of the GH2 exo-beta-mannanase from Xanthomonas axonopodis pv. citri [Xanthomonas citri pv. citri str. 306] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 0.0 | 132 | 1042 | 24 | 921 | Beta-mannosidase A OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=mndA PE=3 SV=1 |
|
| 0.0 | 132 | 1042 | 23 | 909 | Beta-mannosidase A OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=mndA PE=3 SV=1 |
|
| 0.0 | 132 | 1042 | 23 | 909 | Beta-mannosidase A OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=mndA PE=3 SV=1 |
|
| 0.0 | 129 | 1042 | 22 | 927 | Beta-mannosidase A OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=mndA PE=3 SV=2 |
|
| 0.0 | 134 | 1042 | 25 | 923 | Beta-mannosidase A OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=mndA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
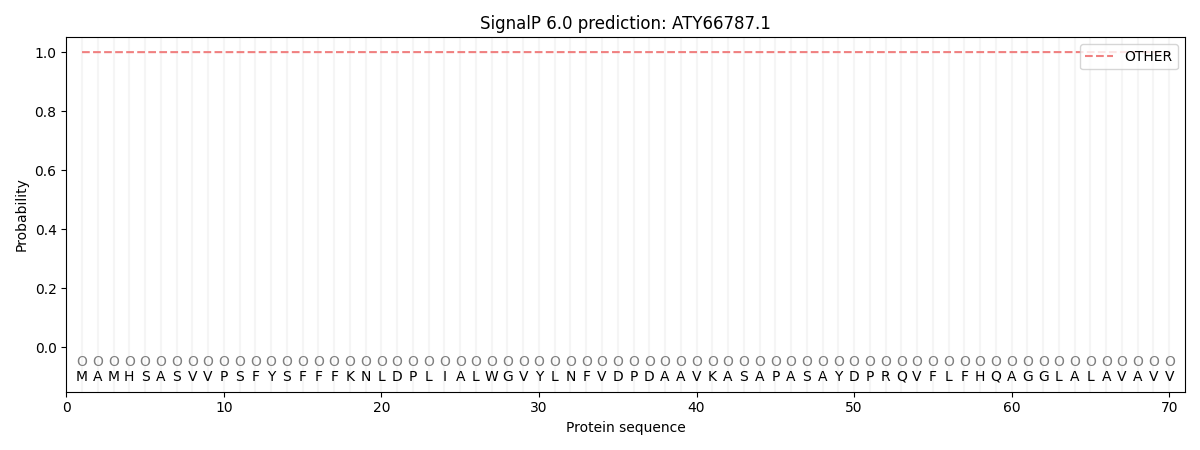
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.999900 | 0.000106 |