You are browsing environment: FUNGIDB
CAZyme Information: ATY66370.1
You are here: Home > Sequence: ATY66370.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Cordyceps militaris | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Cordycipitaceae; Cordyceps; Cordyceps militaris | |||||||||||
| CAZyme ID | ATY66370.1 | |||||||||||
| CAZy Family | GT90 | |||||||||||
| CAZyme Description | AMPK1_CBM domain-containing protein [Source:UniProtKB/TrEMBL;Acc:A0A2H4STE1] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 199889 | E_set_AMPKbeta_like_N | 3.89e-27 | 3 | 81 | 1 | 79 | N-terminal Early set domain, a glycogen binding domain, associated with the catalytic domain of AMP-activated protein kinase beta subunit. E or "early" set domains are associated with the catalytic domain of AMP-activated protein kinase beta subunit glycogen binding domain at the N-terminal end. AMPK is a metabolic stress sensing protein that senses AMP/ATP and has recently been found to act as a glycogen sensor as well. The protein functions as an alpha-beta-gamma heterotrimer. This N-terminal domain is the glycogen binding domain of the beta subunit. This domain is also a member of the CBM48 (Carbohydrate Binding Module 48) family whose members include pullulanase, maltooligosyl trehalose synthase, starch branching enzyme, glycogen branching enzyme, glycogen debranching enzyme, and isoamylase. |
| 406865 | AMPK1_CBM | 1.52e-25 | 5 | 81 | 4 | 78 | Glycogen recognition site of AMP-activated protein kinase. AMPK1_CBM is a family found in close association with AMPKBI pfam04739. The surface of AMPK1_CBM reveals a carbohydrate-binding pocket. |
| 199878 | E_set | 5.36e-07 | 5 | 80 | 3 | 81 | Early set domain associated with the catalytic domain of sugar utilizing enzymes at either the N or C terminus. The E or "early" set domains of sugar utilizing enzymes are associated with different types of catalytic domains at either the N-terminal or C-terminal end. These domains may be related to the immunoglobulin and/or fibronectin type III superfamilies. Members of this family include alpha amylase, sialidase, galactose oxidase, cellulase, cellulose, hyaluronate lyase, chitobiase, and chitinase. A subset of these members were recently identified as members of the CBM48 (Carbohydrate Binding Module 48) family. Members of the CBM48 family include pullulanase, maltooligosyl trehalose synthase, starch branching enzyme, glycogen branching enzyme, glycogen debranching enzyme, isoamylase, and the beta subunit of AMP-activated protein kinase. |
| 237057 | PRK12323 | 1.53e-06 | 489 | 666 | 371 | 580 | DNA polymerase III subunit gamma/tau. |
| 223021 | PHA03247 | 1.90e-06 | 491 | 662 | 2675 | 2840 | large tegument protein UL36; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 708 | 1 | 708 | |
| 2.88e-105 | 1 | 493 | 1 | 508 | |
| 7.74e-105 | 1 | 493 | 170 | 677 | |
| 6.44e-103 | 1 | 493 | 1 | 508 | |
| 9.18e-103 | 1 | 493 | 1 | 523 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7.70e-08 | 6 | 80 | 15 | 86 | the glycogen-binding domain of the AMP-activated protein kinase beta1 subunit [Rattus norvegicus],1Z0M_B the glycogen-binding domain of the AMP-activated protein kinase beta1 subunit [Rattus norvegicus],1Z0M_C the glycogen-binding domain of the AMP-activated protein kinase beta1 subunit [Rattus norvegicus] |
|
| 9.03e-08 | 7 | 80 | 16 | 87 | beta2 carbohydrate binding module (CBM) of AMP-activated protein kinase (AMPK) [Rattus norvegicus],4Y0G_B beta2 carbohydrate binding module (CBM) of AMP-activated protein kinase (AMPK) [Rattus norvegicus],4YEE_A beta2 carbohydrate binding module (CBM) of AMP-activated protein kinase (AMPK) in complex with glucosyl-beta-cyclodextrin [Rattus norvegicus],4YEE_B beta2 carbohydrate binding module (CBM) of AMP-activated protein kinase (AMPK) in complex with glucosyl-beta-cyclodextrin [Rattus norvegicus],4YEE_C beta2 carbohydrate binding module (CBM) of AMP-activated protein kinase (AMPK) in complex with glucosyl-beta-cyclodextrin [Rattus norvegicus],4YEE_D beta2 carbohydrate binding module (CBM) of AMP-activated protein kinase (AMPK) in complex with glucosyl-beta-cyclodextrin [Rattus norvegicus],4YEE_E beta2 carbohydrate binding module (CBM) of AMP-activated protein kinase (AMPK) in complex with glucosyl-beta-cyclodextrin [Rattus norvegicus],4YEE_F beta2 carbohydrate binding module (CBM) of AMP-activated protein kinase (AMPK) in complex with glucosyl-beta-cyclodextrin [Rattus norvegicus],4YEE_G beta2 carbohydrate binding module (CBM) of AMP-activated protein kinase (AMPK) in complex with glucosyl-beta-cyclodextrin [Rattus norvegicus],4YEE_H beta2 carbohydrate binding module (CBM) of AMP-activated protein kinase (AMPK) in complex with glucosyl-beta-cyclodextrin [Rattus norvegicus],4YEE_I beta2 carbohydrate binding module (CBM) of AMP-activated protein kinase (AMPK) in complex with glucosyl-beta-cyclodextrin [Rattus norvegicus],4YEE_J beta2 carbohydrate binding module (CBM) of AMP-activated protein kinase (AMPK) in complex with glucosyl-beta-cyclodextrin [Rattus norvegicus],4YEE_K beta2 carbohydrate binding module (CBM) of AMP-activated protein kinase (AMPK) in complex with glucosyl-beta-cyclodextrin [Rattus norvegicus],4YEE_L beta2 carbohydrate binding module (CBM) of AMP-activated protein kinase (AMPK) in complex with glucosyl-beta-cyclodextrin [Rattus norvegicus],4YEE_M beta2 carbohydrate binding module (CBM) of AMP-activated protein kinase (AMPK) in complex with glucosyl-beta-cyclodextrin [Rattus norvegicus],4YEE_N beta2 carbohydrate binding module (CBM) of AMP-activated protein kinase (AMPK) in complex with glucosyl-beta-cyclodextrin [Rattus norvegicus],4YEE_O beta2 carbohydrate binding module (CBM) of AMP-activated protein kinase (AMPK) in complex with glucosyl-beta-cyclodextrin [Rattus norvegicus],4YEE_P beta2 carbohydrate binding module (CBM) of AMP-activated protein kinase (AMPK) in complex with glucosyl-beta-cyclodextrin [Rattus norvegicus],4YEE_Q beta2 carbohydrate binding module (CBM) of AMP-activated protein kinase (AMPK) in complex with glucosyl-beta-cyclodextrin [Rattus norvegicus],4YEE_R beta2 carbohydrate binding module (CBM) of AMP-activated protein kinase (AMPK) in complex with glucosyl-beta-cyclodextrin [Rattus norvegicus] |
|
| 1.05e-07 | 7 | 80 | 15 | 86 | Glycogen-Binding Domain Of The Amp-Activated Protein Kinase beta2 Subunit [Homo sapiens] |
|
| 1.31e-07 | 7 | 80 | 23 | 94 | Solution NMR structure of the apo-form of the beta2 carbohydrate module of AMP-activated protein kinase [Rattus norvegicus],2LU4_A Solution NMR structure of the beta2 carbohydrate module of AMP-activated protein kinase bound to glucosyl-cyclodextrin [Rattus norvegicus] |
|
| 1.96e-07 | 6 | 80 | 15 | 86 | Chain A, 5'-AMP-activated protein kinase, beta-1 subunit [Rattus norvegicus],1Z0N_B Chain B, 5'-AMP-activated protein kinase, beta-1 subunit [Rattus norvegicus],1Z0N_C Chain C, 5'-AMP-activated protein kinase, beta-1 subunit [Rattus norvegicus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.88e-09 | 4 | 165 | 8 | 148 | Signal transduction protein MDG1 OS=Saccharomyces cerevisiae (strain YJM789) OX=307796 GN=MDG1 PE=3 SV=1 |
|
| 9.54e-09 | 1 | 90 | 6 | 105 | Cruciform DNA-recognizing protein 1 OS=Saccharomyces cerevisiae (strain VIN 13) OX=764099 GN=CRP1 PE=3 SV=2 |
|
| 9.54e-09 | 1 | 90 | 6 | 105 | Cruciform DNA-recognizing protein 1 OS=Saccharomyces cerevisiae (strain AWRI796) OX=764097 GN=CRP1 PE=3 SV=2 |
|
| 9.54e-09 | 1 | 90 | 6 | 105 | Cruciform DNA-recognizing protein 1 OS=Saccharomyces cerevisiae (strain Lalvin QA23) OX=764098 GN=CRP1 PE=3 SV=2 |
|
| 9.54e-09 | 1 | 90 | 6 | 105 | Cruciform DNA-recognizing protein 1 OS=Saccharomyces cerevisiae (strain YJM789) OX=307796 GN=CRP1 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
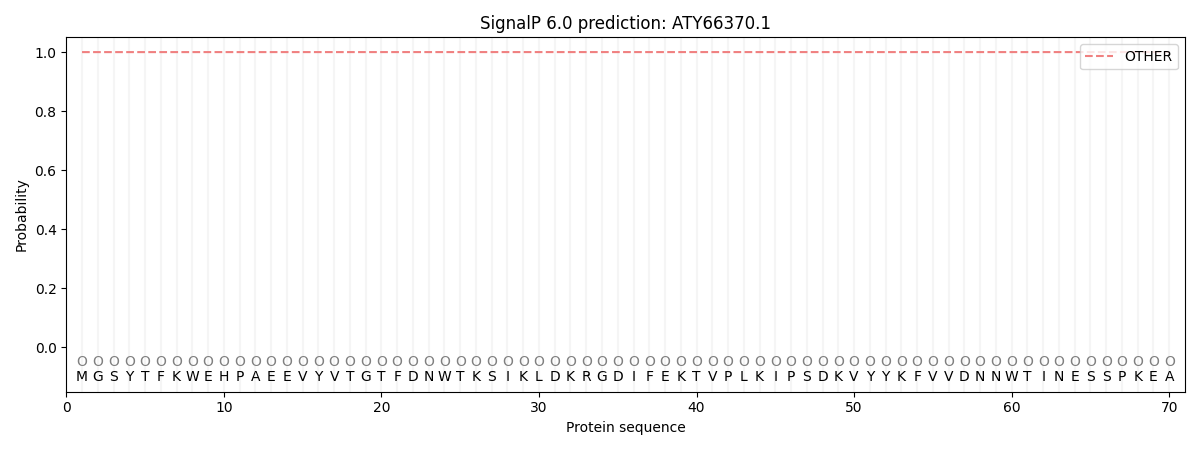
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000046 | 0.000000 |
