You are browsing environment: FUNGIDB
CAZyme Information: ATY66193.1
You are here: Home > Sequence: ATY66193.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Cordyceps militaris | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Cordycipitaceae; Cordyceps; Cordyceps militaris | |||||||||||
| CAZyme ID | ATY66193.1 | |||||||||||
| CAZy Family | GT2 | |||||||||||
| CAZyme Description | beta-fructofuranosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.26:5 | 3.2.1.80:2 | 2.4.1.-:1 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH32 | 60 | 375 | 9.6e-88 | 0.9829351535836177 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 350134 | GH32_Inu-like | 1.25e-137 | 65 | 362 | 1 | 286 | glycoside hydrolase family 32 protein such as Aspergillus ficuum endo-inulinase (Inu2). This subfamily of glycosyl hydrolase family GH32 includes endo-inulinase (inu2, EC 3.2.1.7), exo-inulinase (Inu1, EC 3.2.1.80), invertase (EC 3.2.1.26), and levan fructotransferase (LftA, EC 4.2.2.16), among others. These enzymes cleave sucrose into fructose and glucose via beta-fructofuranosidase activity, producing invert sugar that is a mixture of dextrorotatory D-glucose and levorotatory D-fructose, thus named invertase (EC 3.2.1.26). These retaining enzymes (i.e. they retain the configuration at anomeric carbon atom of the substrate) catalyze hydrolysis in two steps involving a covalent glycosyl enzyme intermediate: an aspartate located close to the N-terminus acts as the catalytic nucleophile and a glutamate acts as the general acid/base; a conserved aspartate residue in the Arg-Asp-Pro (RDP) motif stabilizes the transition state. These enzymes are predicted to display a 5-fold beta-propeller fold as found for GH43 and CH68. The breakdown of sucrose is widely used as a carbon or energy source by bacteria, fungi, and plants. Invertase is used commercially in the confectionery industry, since fructose has a sweeter taste than sucrose and a lower tendency to crystallize. A common structural feature of all these enzymes is a 5-bladed beta-propeller domain, similar to GH43, that contains the catalytic acid and catalytic base. A long V-shaped groove, partially enclosed at one end, forms a single extended substrate-binding surface across the face of the propeller. |
| 214757 | Glyco_32 | 2.15e-103 | 60 | 506 | 1 | 437 | Glycosyl hydrolases family 32. |
| 224536 | SacC | 2.86e-103 | 55 | 545 | 28 | 486 | Sucrose-6-phosphate hydrolase SacC, GH32 family [Carbohydrate transport and metabolism]. |
| 395193 | Glyco_hydro_32N | 9.17e-89 | 60 | 361 | 1 | 293 | Glycosyl hydrolases family 32 N-terminal domain. This domain corresponds to the N-terminal domain of glycosyl hydrolase family 32 which forms a five bladed beta propeller structure. |
| 350110 | GH32_FFase | 1.23e-68 | 66 | 362 | 1 | 278 | Glycosyl hydrolase family 32, beta-fructosidases. Glycosyl hydrolase family GH32 cleaves sucrose into fructose and glucose via beta-fructofuranosidase activity, producing invert sugar that is a mixture of dextrorotatory D-glucose and levorotatory D-fructose, thus named invertase (EC 3.2.1.26). This family also contains other fructofuranosidases such as inulinase (EC 3.2.1.7), exo-inulinase (EC 3.2.1.80), levanase (EC 3.2.1.65), and transfructosidases such sucrose:sucrose 1-fructosyltransferase (EC 2.4.1.99), fructan:fructan 1-fructosyltransferase (EC 2.4.1.100), sucrose:fructan 6-fructosyltransferase (EC 2.4.1.10), fructan:fructan 6G-fructosyltransferase (EC 2.4.1.243) and levan fructosyltransferases (EC 2.4.1.-). These retaining enzymes (i.e. they retain the configuration at anomeric carbon atom of the substrate) catalyze hydrolysis in two steps involving a covalent glycosyl enzyme intermediate: an aspartate located close to the N-terminus acts as the catalytic nucleophile and a glutamate acts as the general acid/base; a conserved aspartate residue in the Arg-Asp-Pro (RDP) motif stabilizes the transition state. These enzymes are predicted to display a 5-fold beta-propeller fold as found for GH43 and CH68. The breakdown of sucrose is widely used as a carbon or energy source by bacteria, fungi, and plants. Invertase is used commercially in the confectionery industry, since fructose has a sweeter taste than sucrose and a lower tendency to crystallize. A common structural feature of all these enzymes is a 5-bladed beta-propeller domain, similar to GH43, that contains the catalytic acid and catalytic base. A long V-shaped groove, partially enclosed at one end, forms a single extended substrate-binding surface across the face of the propeller. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 580 | 1 | 580 | |
| 3.94e-301 | 9 | 575 | 12 | 569 | |
| 4.76e-298 | 16 | 569 | 9 | 556 | |
| 3.17e-281 | 37 | 575 | 31 | 568 | |
| 7.84e-281 | 41 | 563 | 29 | 545 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.47e-110 | 56 | 535 | 8 | 498 | Structure of Saccharomyces cerevisiae invertase [Saccharomyces cerevisiae S288C],4EQV_B Structure of Saccharomyces cerevisiae invertase [Saccharomyces cerevisiae S288C],4EQV_C Structure of Saccharomyces cerevisiae invertase [Saccharomyces cerevisiae S288C],4EQV_D Structure of Saccharomyces cerevisiae invertase [Saccharomyces cerevisiae S288C],4EQV_E Structure of Saccharomyces cerevisiae invertase [Saccharomyces cerevisiae S288C],4EQV_F Structure of Saccharomyces cerevisiae invertase [Saccharomyces cerevisiae S288C],4EQV_G Structure of Saccharomyces cerevisiae invertase [Saccharomyces cerevisiae S288C],4EQV_H Structure of Saccharomyces cerevisiae invertase [Saccharomyces cerevisiae S288C] |
|
| 9.61e-98 | 56 | 518 | 39 | 528 | The crystal structure of exoinulinase INU1 [Kluyveromyces marxianus DMKU3-1042],6J0T_B The crystal structure of exoinulinase INU1 [Kluyveromyces marxianus DMKU3-1042] |
|
| 4.36e-92 | 56 | 515 | 10 | 476 | Chain A, Invertase [Schwanniomyces occidentalis],3KF3_B Chain B, Invertase [Schwanniomyces occidentalis] |
|
| 4.74e-92 | 56 | 515 | 13 | 479 | Chain A, Invertase [Schwanniomyces occidentalis],3KF5_B Chain B, Invertase [Schwanniomyces occidentalis] |
|
| 6.84e-91 | 56 | 515 | 36 | 502 | Chain A, Fructofuranosidase [Schwanniomyces occidentalis],3U75_B Chain B, Fructofuranosidase [Schwanniomyces occidentalis],3U75_C Chain C, Fructofuranosidase [Schwanniomyces occidentalis],3U75_D Chain D, Fructofuranosidase [Schwanniomyces occidentalis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.22e-109 | 56 | 535 | 28 | 518 | Invertase 2 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=SUC2 PE=1 SV=1 |
|
| 5.02e-108 | 56 | 535 | 28 | 518 | Invertase 1 OS=Saccharomyces cerevisiae OX=4932 GN=SUC1 PE=1 SV=1 |
|
| 5.54e-107 | 56 | 535 | 28 | 518 | Invertase 4 OS=Saccharomyces cerevisiae OX=4932 GN=SUC4 PE=3 SV=1 |
|
| 7.06e-99 | 56 | 529 | 20 | 495 | Extracellular exo-inulinase OS=Meyerozyma guilliermondii (strain ATCC 6260 / CBS 566 / DSM 6381 / JCM 1539 / NBRC 10279 / NRRL Y-324) OX=294746 GN=PGUG_02777 PE=1 SV=2 |
|
| 7.74e-98 | 56 | 529 | 20 | 495 | Extracellular exo-inulinase inuE OS=Meyerozyma guilliermondii OX=4929 PE=1 SV=3 |
SignalP and Lipop Annotations help
This protein is predicted as SP
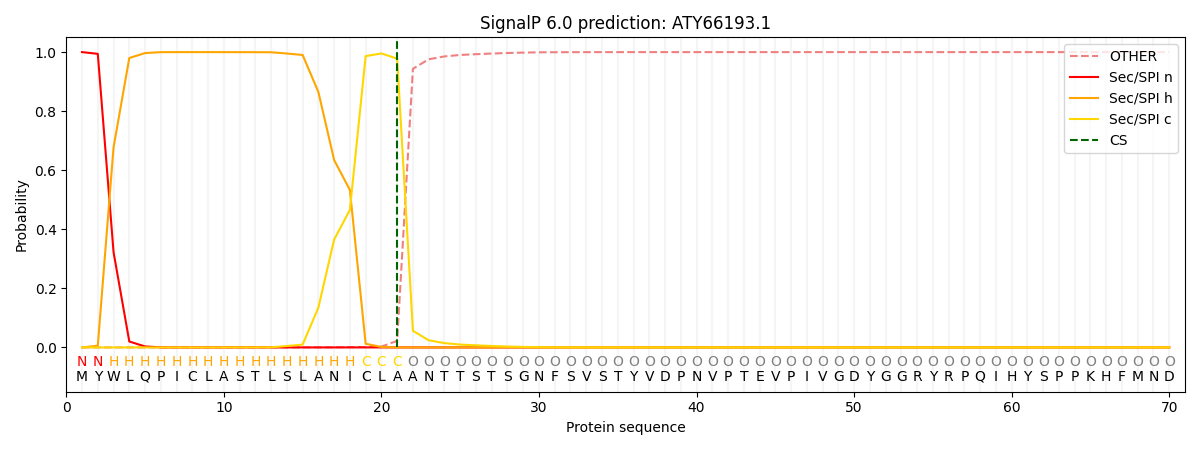
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000292 | 0.999687 | CS pos: 21-22. Pr: 0.9776 |
